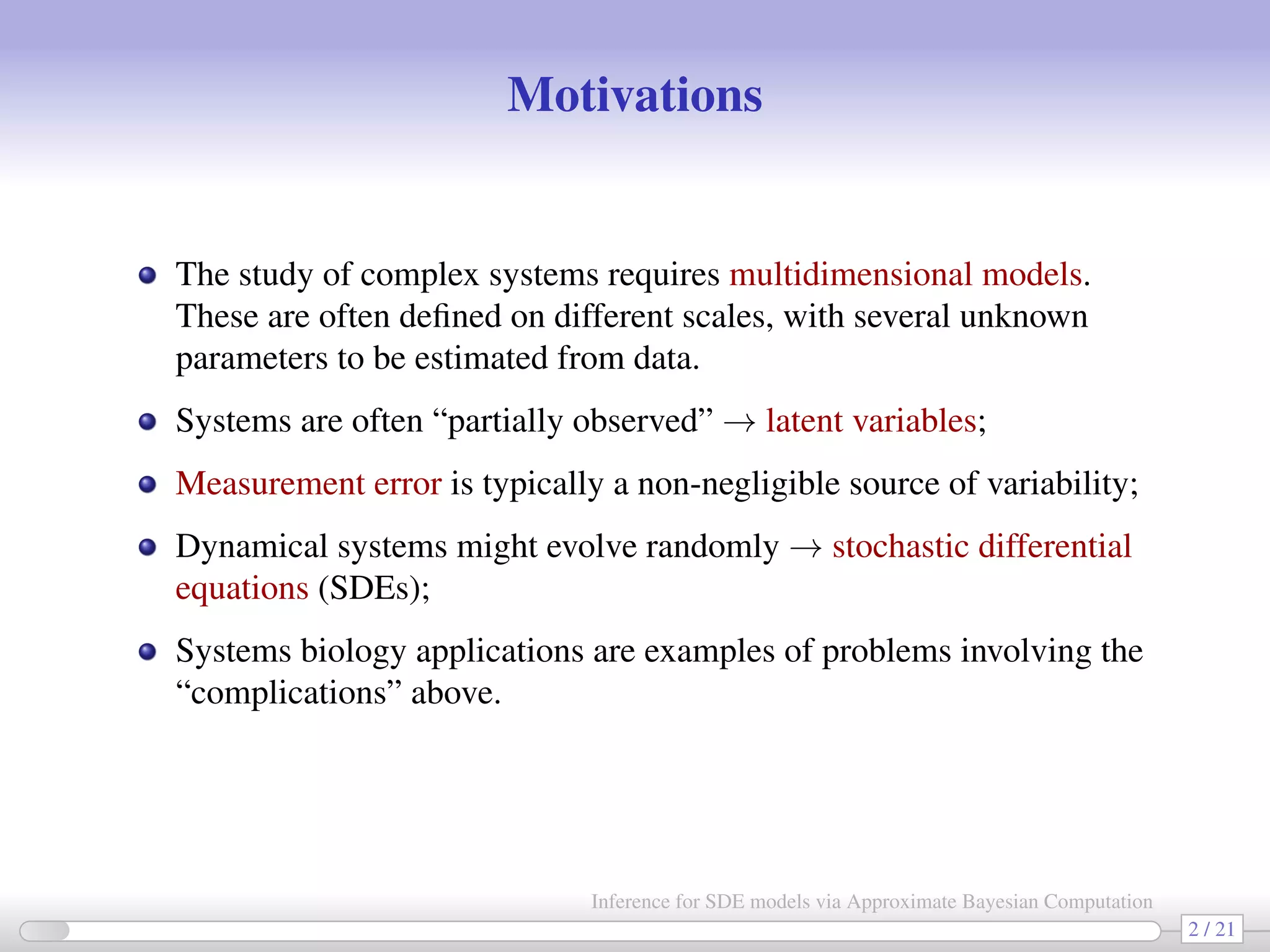
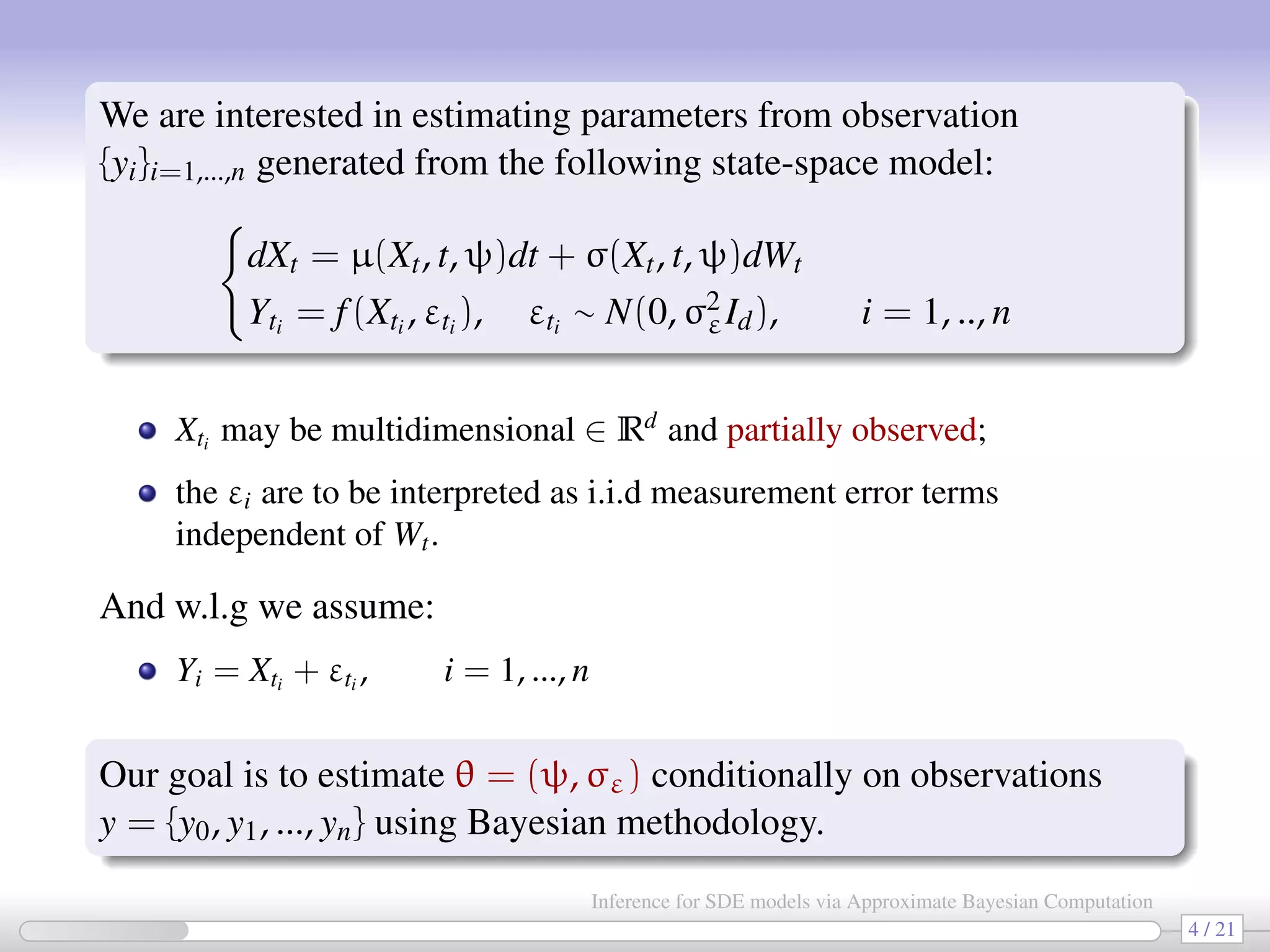
The document discusses the inference for stochastic differential equation (SDE) models using approximate Bayesian computation (ABC) methodologies. It highlights the challenges posed by complex parameter estimation in partially observed systems and proposes ABC as a viable alternative to traditional MCMC methods, particularly for large systems where likelihood computations are unfeasible. The document includes various algorithms and examples illustrating the implementation of ABC in estimating parameters for SDE models in applications such as systems biology and stochastic kinetic networks.


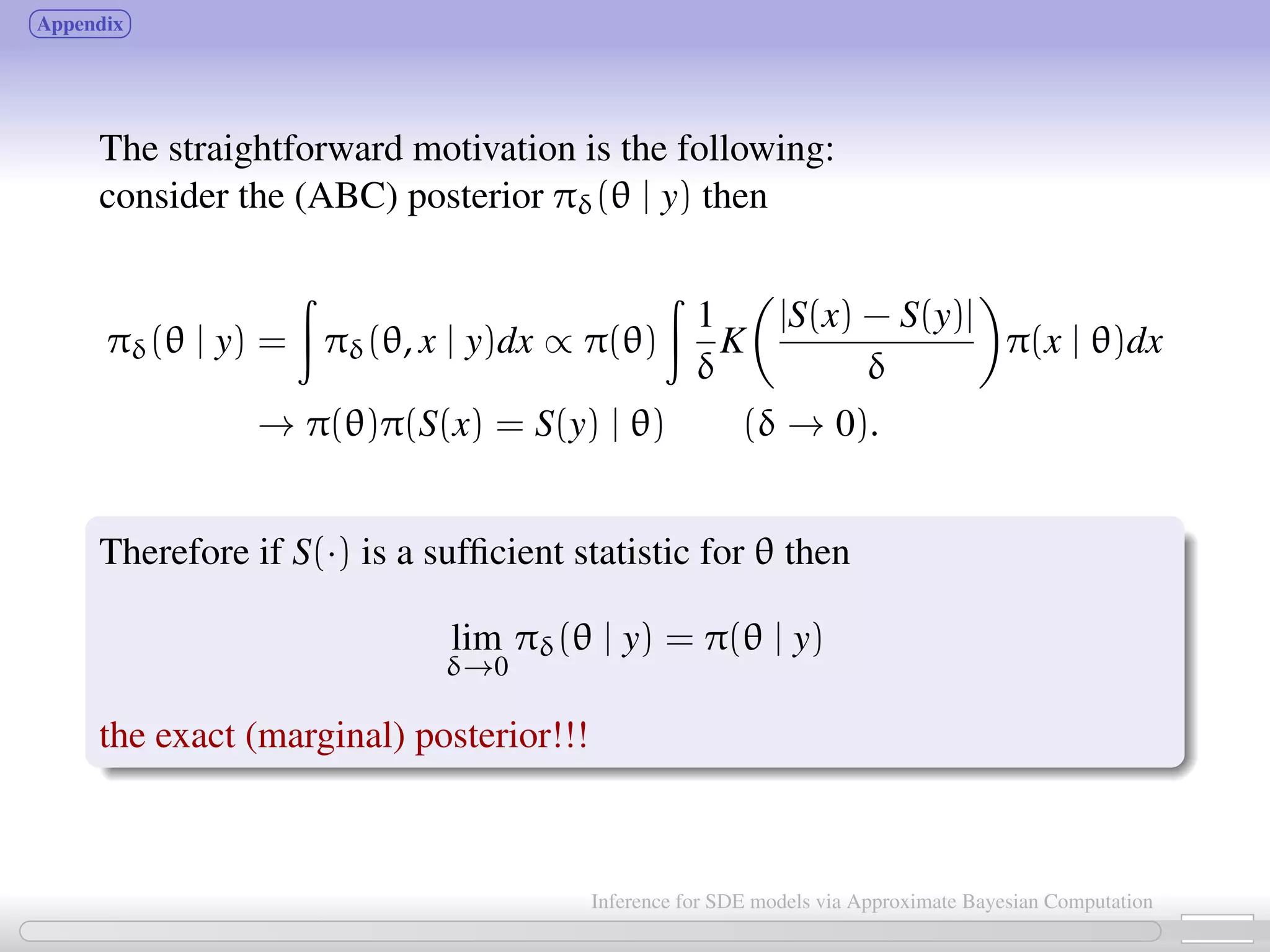
![Bayesian inference about θ is based on the (marginal) posterior density π(θ | y) ∝ l(θ; y)π(θ) We assume that the likelihood function l(θ; y) may be unavailable for mathematical reasons (not available in closed from) or for computational reasons (too expensive to calculate). In our case data: y = (y0, y1, ..., yn) obtained at times {t0, ..., tn} i.e. yi ≡ yti and x = (x0, x1, ..., xn) is the SDE solution at time-points {t0, ..., tn}. l(θ; y) = π(y | θ) = π(y | x; θ)π(x | θ)dx → “data augmentation” from θ to (x, θ) use MCMC to deal with the multiple integration problem. Because of increased dimension (from θ to (θ, x)) convergence properties of MCMC algorithms are too poor for the algorithm to be considered. [Marin, Pudlo, Robert and Ryder 2011] 5 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-5-2048.jpg)
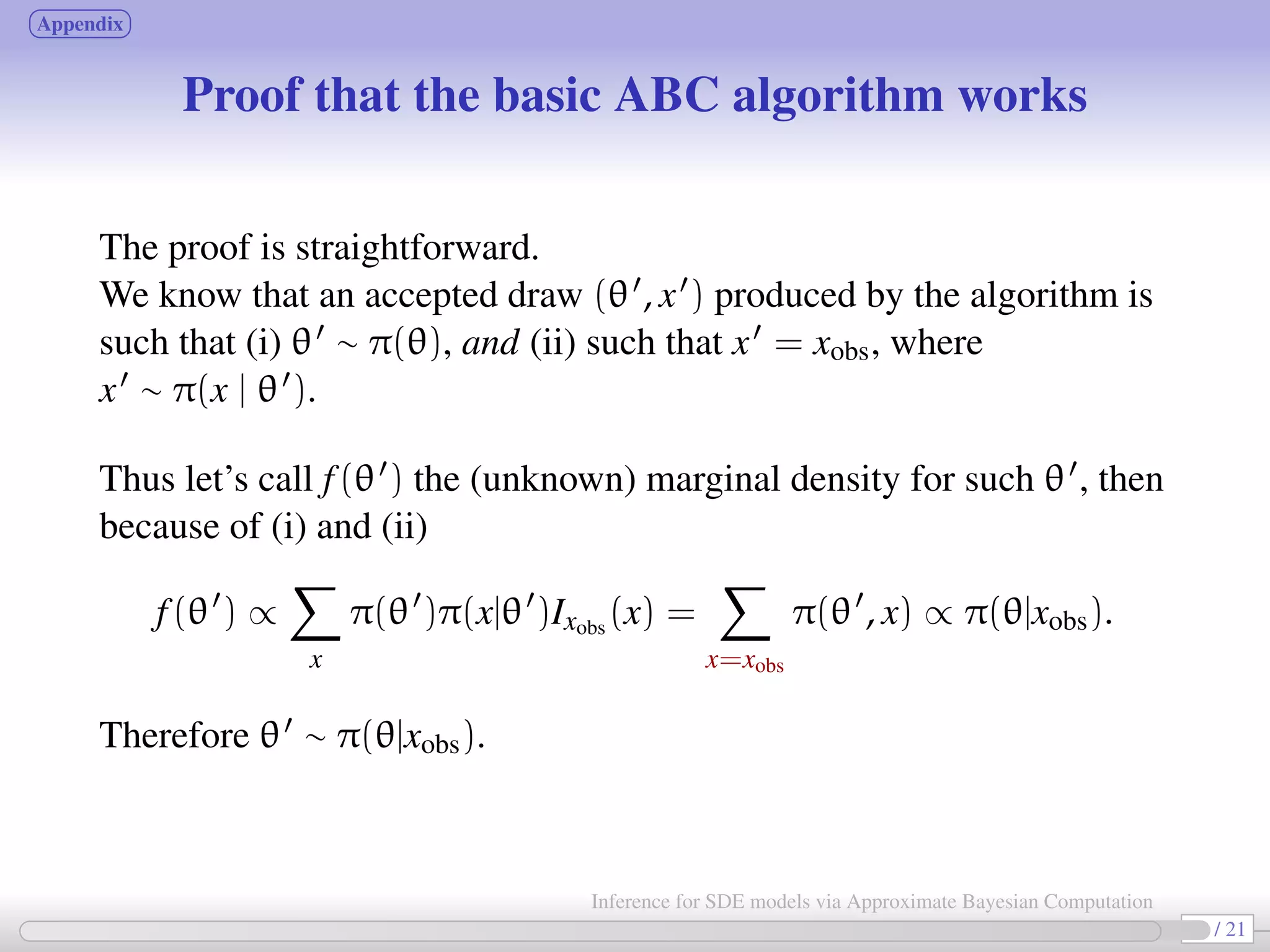
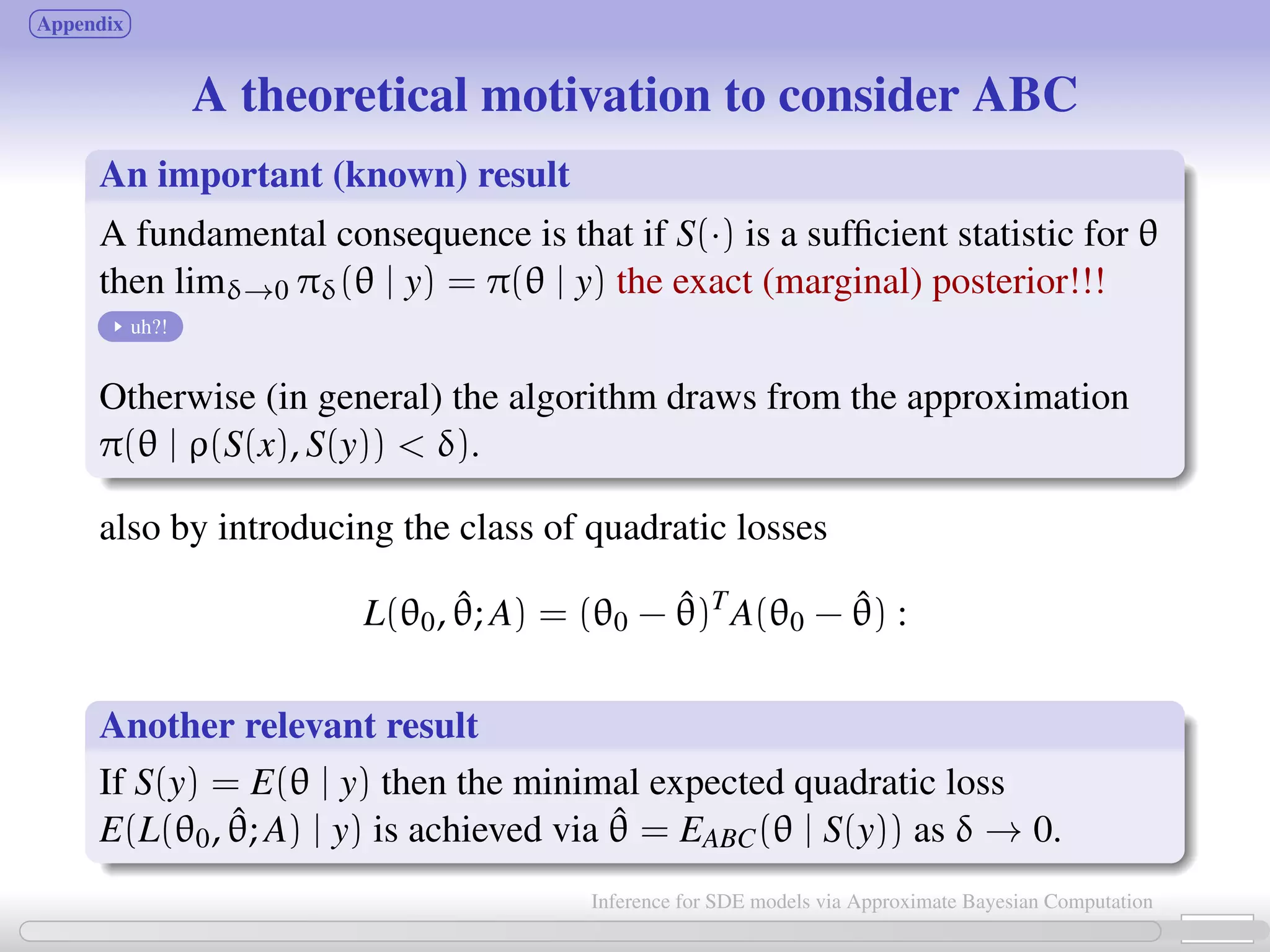
![Approximate Bayesian Computation (ABC, first proposed in Tavaré et al., 1997) bypass the computation of the likelihood function. Basic ABC idea (for sake of simplicity: no measurement error here): for an observation xobs ∼ π(x | θ), under the prior π(θ), keep jointly simulating θ ∼ π(θ), x ∼ π(x | θ ) until the simulated variable x is equal to the observed one, x = xobs. Then a θ satisfying such condition is such that θ ∼ π(θ | xobs). [Tavaré et al., 1997]. proof The equality x = xobs can’t usually be expected for large discrete systems, and holds with zero probability for vectors x = (xt0 , ..., xtn ) resulting from a diffusion process. Choose instead an accuracy threshold δ; substitute “accept if x = xobs” with “accept if ρ(S(x ), S(xobs)) < δ” for some measure ρ(·) and statistic S(·). 6 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-6-2048.jpg)
![As from the previous slide, for dynamical problems we only need to know how to generate a proposal x = (x0, ..., xn) ⇒ for SDEs: use Euler-Maruyama, Milstein, Stochastic RK discretizations etc. The simplest approximation scheme to the SDE solution (Euler–Maruyama): given an SDE: dXt = µ(Xt, ψ)dt + σ(Xt, ψ)dWt approximate with Xt = Xt−h + µ(Xt−h, ψ)h + σ(Xt−h, ψ)(Wt − Wt−h) [approximation converge to the exact solution for h → 0] and we can interpolate on such approximation at discrete times {t0, t1, ..., tn} to obtain the corresponding “latent states” x = {x0, x1, ..., xn}. 8 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-8-2048.jpg)
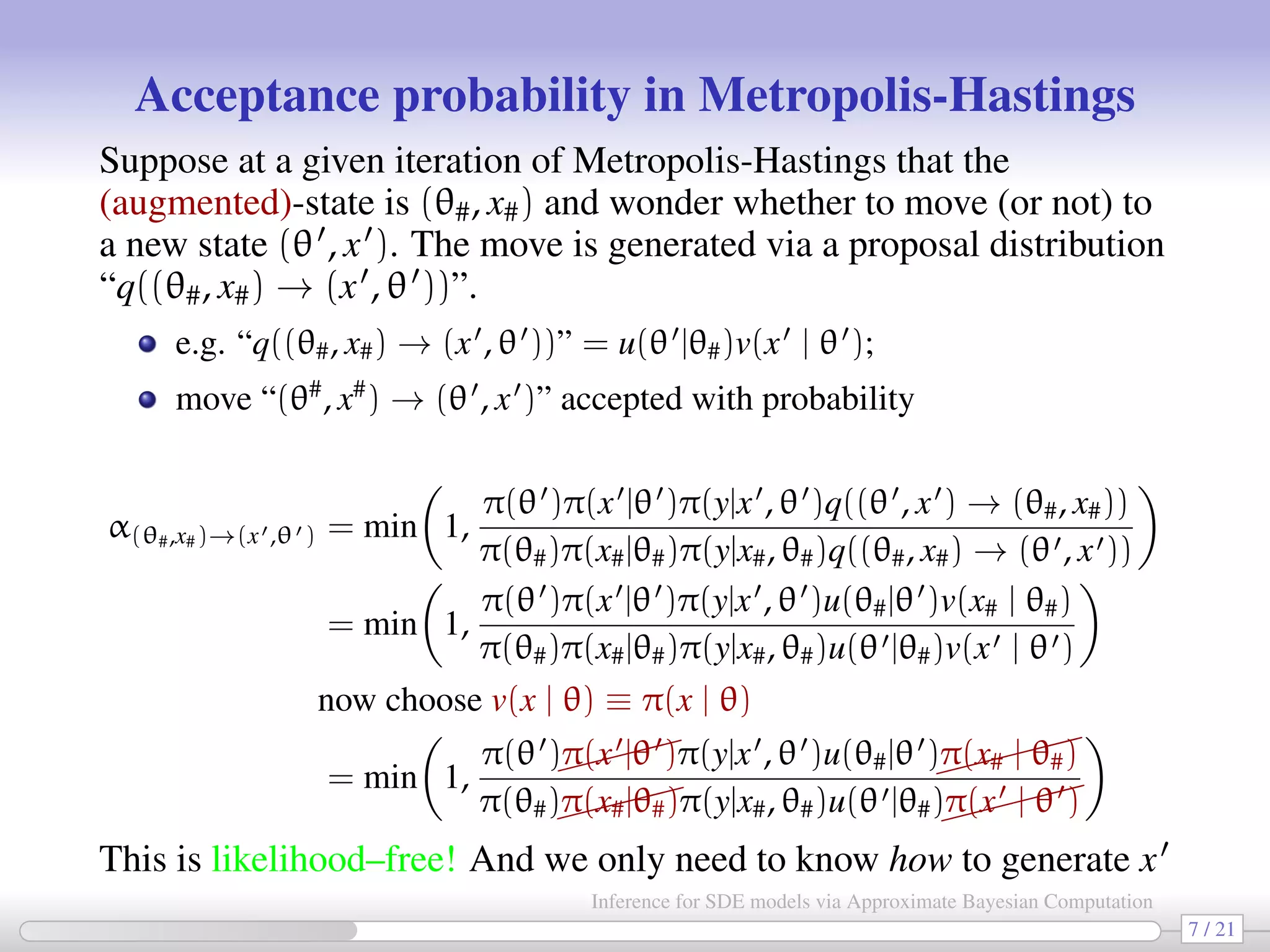
![We now plug the previous ideas in a MCMC ABC algorithm [Sisson-Fan, 2011] 1. Choose or simulate θstart ∼ π(θ) and xstart ∼ usually unknown! π(x|θstart) . Fix δ 0 and r = 0. Put θr = θstart and (supposedly known!) statistics S(xr) ≡ S(xstart). At (r + 1)th MCMC iteration: 2. generate θ ∼ u(θ |θr) from its proposal distribution; 3. generate x ∼ π(x |θ ) and calculate S(x ); 4. with probability min 1, π(θ )πδ(y|x ,θ ))u(θr|θ ) π(θr)πδ(y|xr,θr)u(θ |θr) set (θr+1, S(xr+1)) = (θ , S(x )) otherwise set (θr+1, S(xr+1)) = (θr, S(xr)); 5. increment r to r + 1 and go to step 2. with e.g. πδ(y | x, θ) = 1 δK |S(x)−S(y)| δ [Bloom 2010]. 9 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-9-2048.jpg)
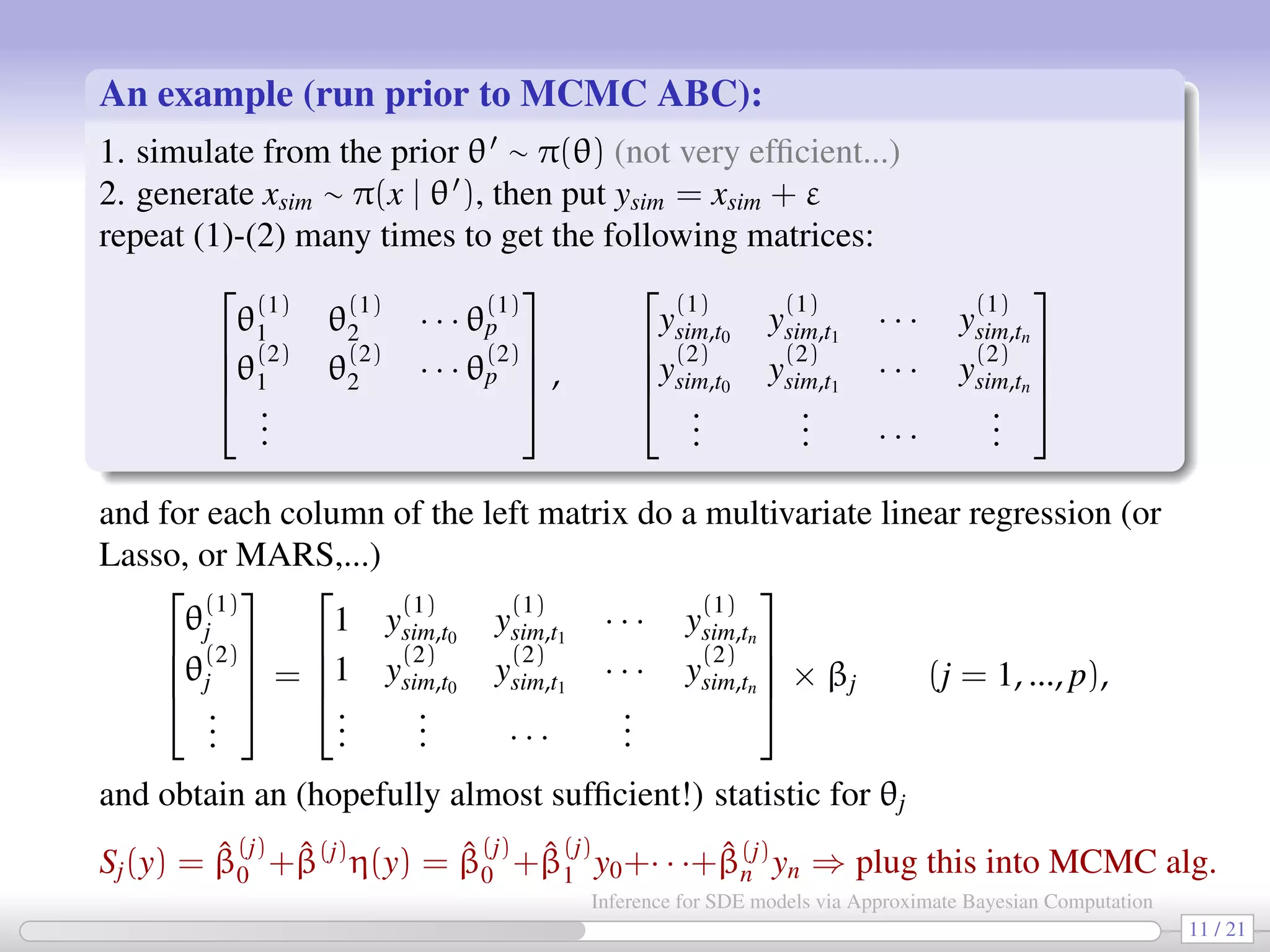
![The previous algorithm generates a sequence {θr, xr} from the (ABC) posterior πδ(θ, xr | y). We only retain the {θr} which are thus from πδ(θ | y), the (marginal) ABC posterior of θ. In order to apply ABC methodology we are required to specify the statistic S(·) for θ. [Fearnhead Prangle 2012] (Classic result of Bayesian stats: for quadratic losses) the “optimal” choice of S(y) is given by S(y) = E(θ | y), the (unknown) posterior of θ. So Fearnhead Prangle propose a regression-based approach to determine S(·) (prior to MCMC start): for the jth parameter in θ fit the following linear regression models Sj(y) = ˆE(θj|y) = ˆβ (j) 0 + ˆβ(j) η(y), j = 1, 2, ..., dim(θ) [e.g. Sj(y) = ˆβ (j) 0 + ˆβ(j) η(y) = ˆβ (j) 0 + ˆβ (j) 1 y0 + · · · + ˆβ (j) n yn] repeat the fitting separately for each θj. hopefully Sj(y) = ˆβ (j) 0 + ˆβ(j) η(y) will be “informative” for θj. 10 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-10-2048.jpg)
![In some cases we have obtained better results (than with linear regression) using statistics based on: regression via MARS (multivariate adaptive regression splines [Friedman 1991]); Lasso-like estimation via glmnet [Friedman, Hastie, Tibshirani 2001]: estimates are found via ˆβ = argminβ 1 2 n i=0 (yi − p j=1 ηijβj)2 + γ p j=1 | βj | and the penality γ selected via cross validation methods (the one giving the smallest mean prediction error is selected). 12 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-12-2048.jpg)
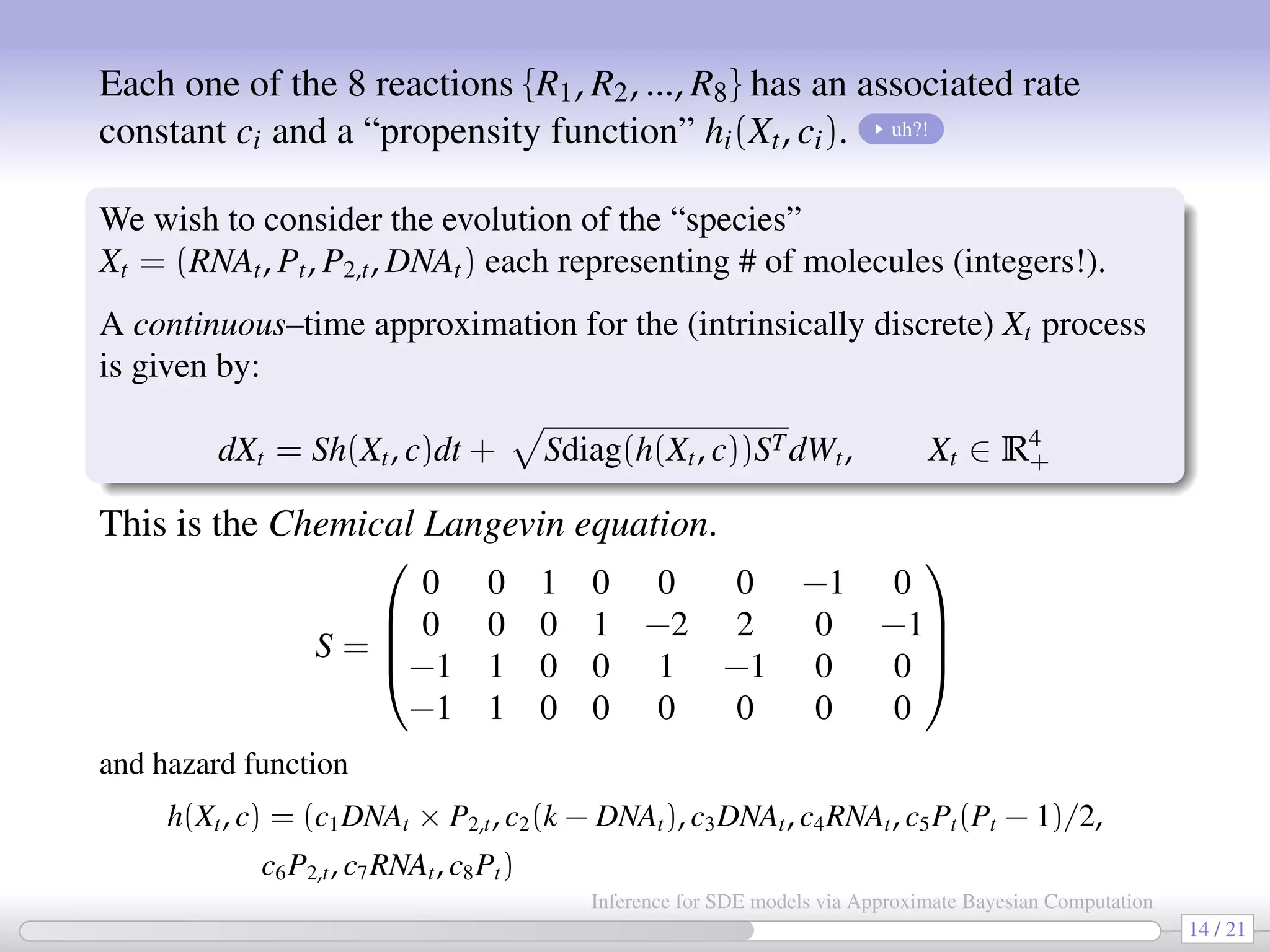
![An application: Stochastic kinetic networks [also in Golightly-Wilkinson, 2010] Consider a set of reactions {R1, R2, ..., R8}: R1 : DNA + P2 → DNA · P2 R2 : DNA · P2 → DNA + P2 R3 : DNA → DNA + RNA R4 : RNA → RNA + P R5 : 2P → P2 (dimerization) R6 : P2 → 2P (dimerization) R7 : RNA → ∅ (degradation) R8 : P → ∅ (degradation) These reactions represent a simplified model for prokaryotic auto-regulation. “Reactants” are on the left side of →; “Products” are on the right side of →; There are several ways to simulate biochemical networks, e.g. the “exact” Gillespie algorithm (computationally intense for inferential purposes). Or by using a diffusion approximation ⇒ SDE. 13 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-13-2048.jpg)
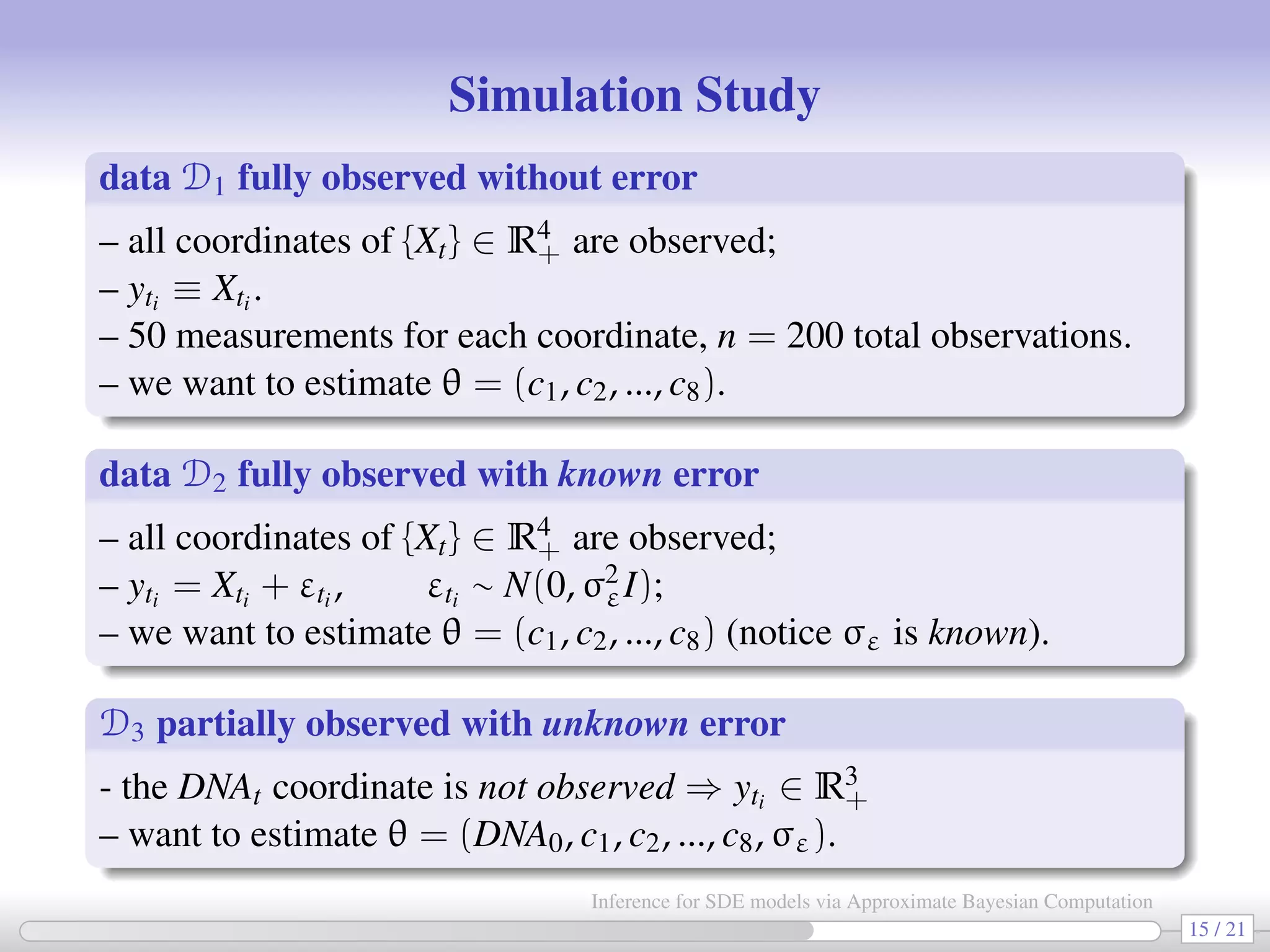
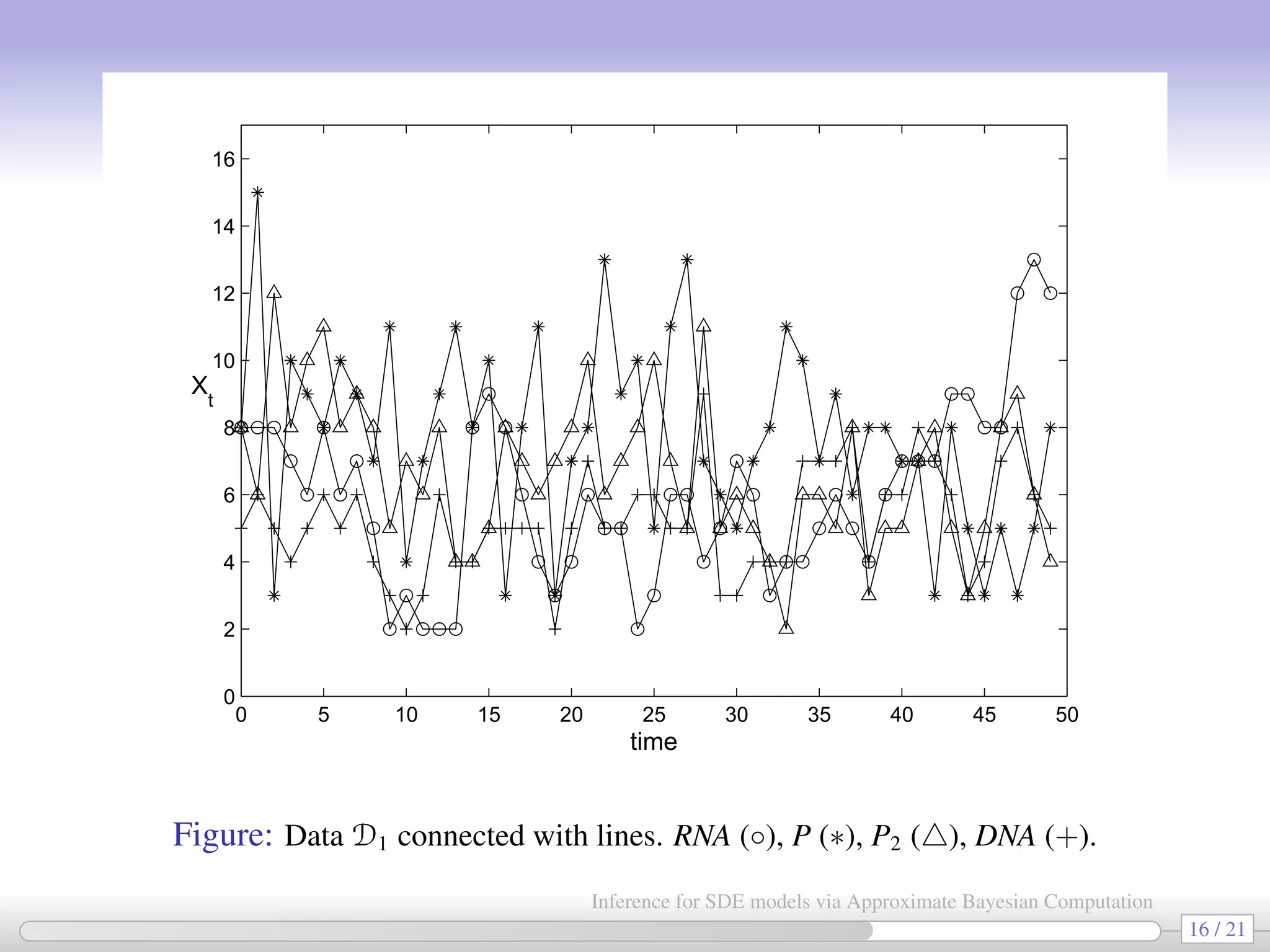
![In all 3 cases a chain having 3,000,000 elements is generated (∼15 hrs with Matlab). Priors: log cj ∼ U(−3, 0) and log DNA0 ∼ U(0, 2.3) (when needed). True values D1 D2 D3 DNA0 5 — — 3.110 [1.441, 6.526] c1 0.1 0.074 0.074 0.074 [0.057, 0.097] [0.055, 0.099] [0.056, 0.097] c2 0.7 0.521 0.552 0.536 [0.282, 0.970] [0.294, 1.029] [0.345, 0.832] c1/c2 0.143 0.142 0.135 0.138 c3 0.35 0.216 0.214 0.244 [0.114, 0.4081] [0.109, 0.417] [0.121, 0.492] c4 0.2 0.164 0.168 0.174 [0.088, 0.308] [0.088, 0.312] [0.089, 0.331] c5 0.1 0.088 0.088 0.087 [0.070, 0.112] [0.069, 0.114] [0.063, 0.120] c6 0.9 0.352 0.355 0.379 [0.139, 0.851] [0.136, 0.880] [0.147, 0.937] c5/c6 0.111 0.250 0.248 0.228 c7 0.3 0.158 0.158 0.157 [0.113, 0.221] [0.108, 0.229] [0.103, 0.240] c8 0.1 0.160 0.169 0.165 [0.098, 0.266] [0.097, 0.286] [0.091, 0.297] σε 1.414 — — 0.652 [0.385, 1.106] 17 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-17-2048.jpg)
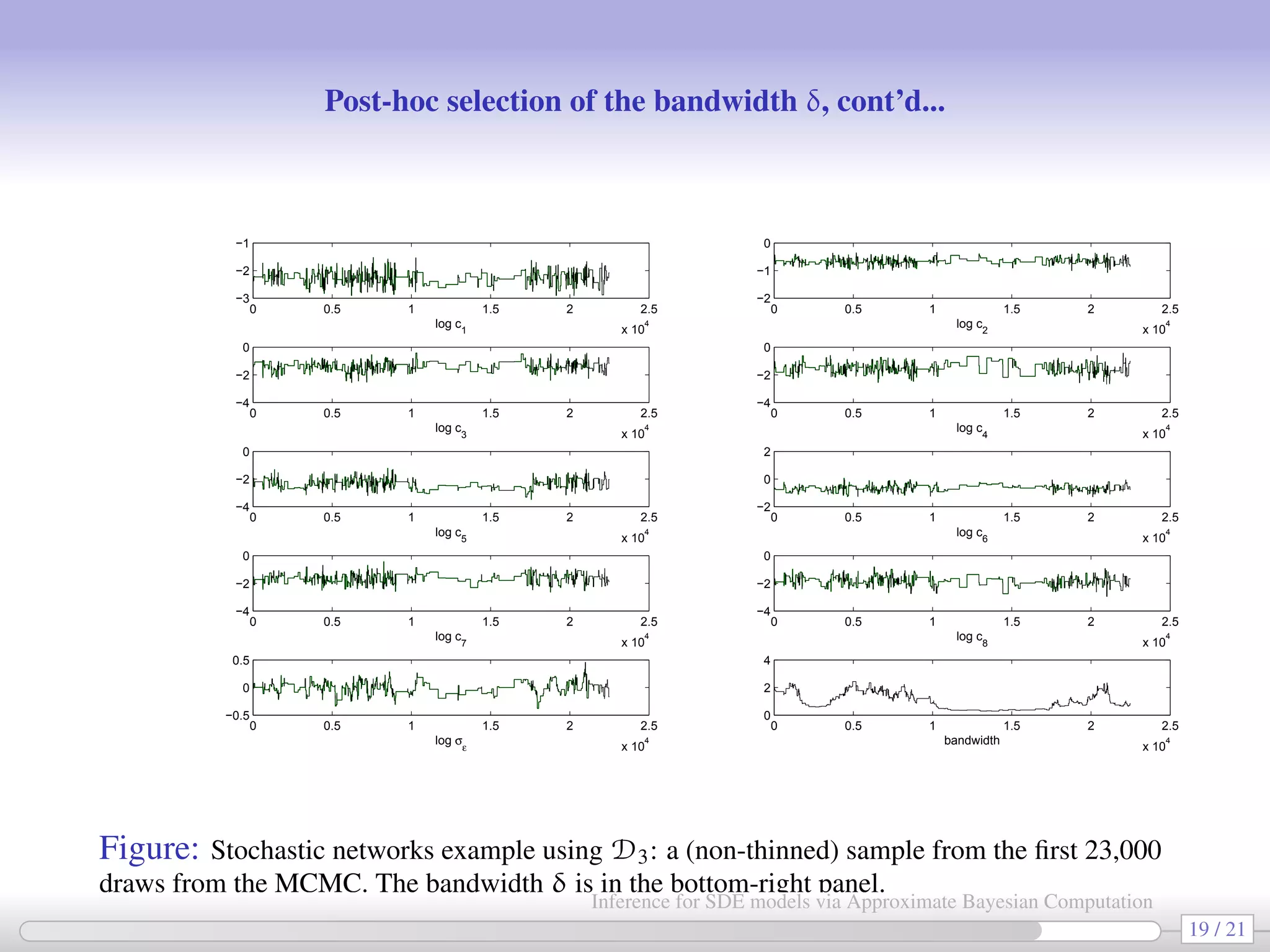
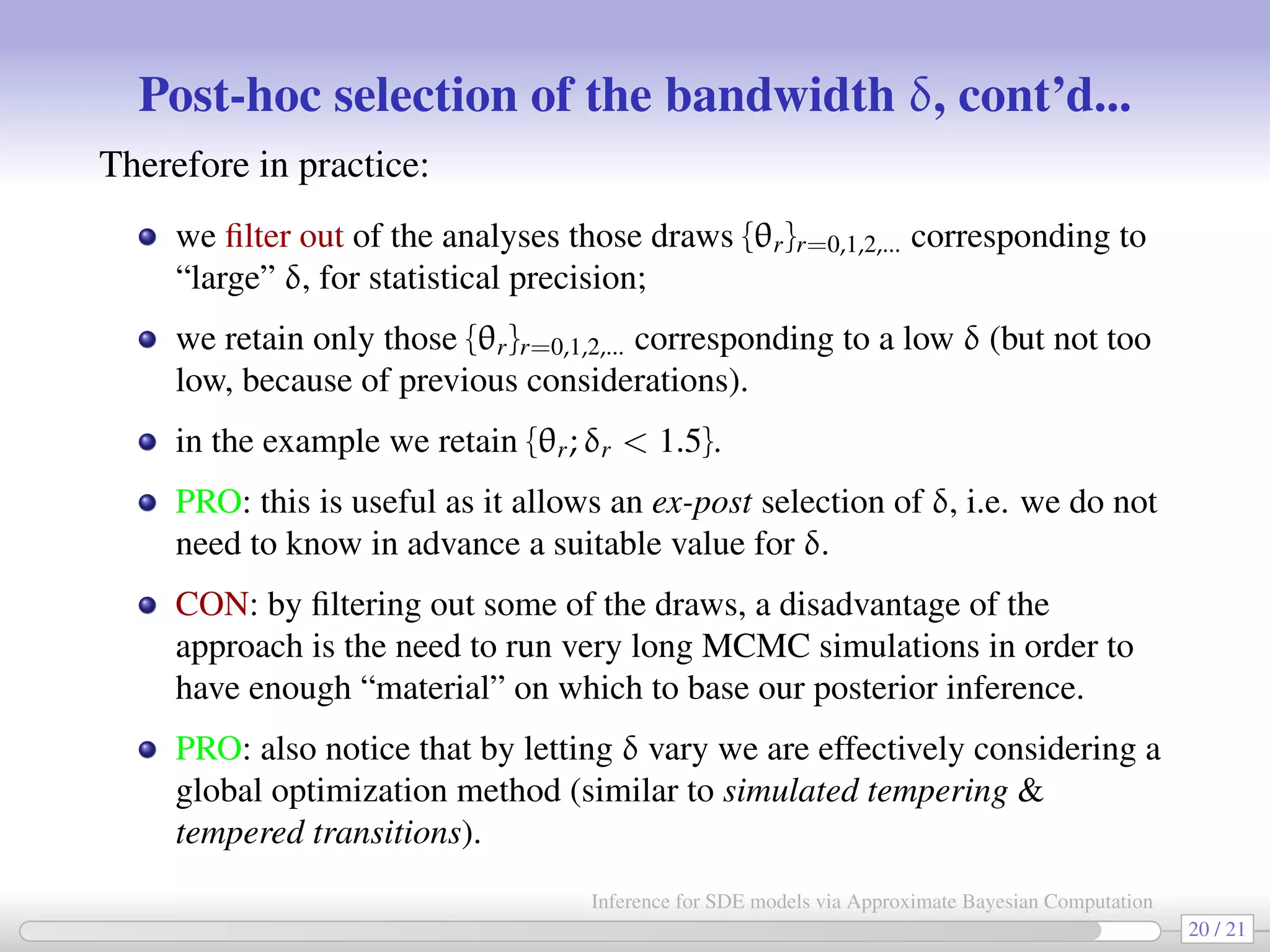
![HOWTO: post-hoc selection of δ (the “precision” parameter) [Bortot et al. 2007] As previously explained, during the MCMC we let δ vary (according to a MRW): at rth iteration δr = δr−1 + ∆, with ∆ ∼ N(0, ν2 ). After the end of the MCMC we have a sequence {θr, δr}r=0,1,2... and for each parameter {θj,r}r=0,1,2... we produce a plot like the following (e.g. log c3 vs δ): 0 0.5 1 1.5 2 2.5 3 3.5 4 −2.5 −2 −1.5 −1 −0.5 0 bandwidth 18 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-18-2048.jpg)
![Appendix Some remarks on SysBio concepts a “propensity function” hi(Xt, ci) gives the overall hazard of a type i reaction occurring, that is the probability of a type i reaction occurring in the time interval (t, t + dt] is hi(Xt, ci)dt. a “conservation law” (from redundant rows in S): DNA · P2 + DNA = k. In a stoichiometry matrix S reactants appear as negative values and products appear as positive values. 25 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-25-2048.jpg)
![Appendix Choice of kernel function We follow Fearnhead-Prangle(2012): we consider bounded kernels K(·) 1; specifically we use the uniform kernel K(z) returning 1 when zT Az c and 0 otherwise. In our case z = (S(ysim) − S(y))/δ and A is chosen to be a p × p diagonal matrix defining the relative weighting of the parameters in the loss function. The uniform kernel is defined on a region zT Az bounded by a volume c, with c = Vp|A|1/p , where Vp = π−1 [Γ(p/2)p/2]2/p and p = dim(θ); such c is the unique value producing a valid probability density function K i.e. such that the volume of the region zT Az c equals 1. 26 / 21 Inference for SDE models via Approximate Bayesian Computation](https://image.slidesharecdn.com/abcumea-160220184210/75/Inference-for-stochastic-differential-equations-via-approximate-Bayesian-computation-26-2048.jpg)