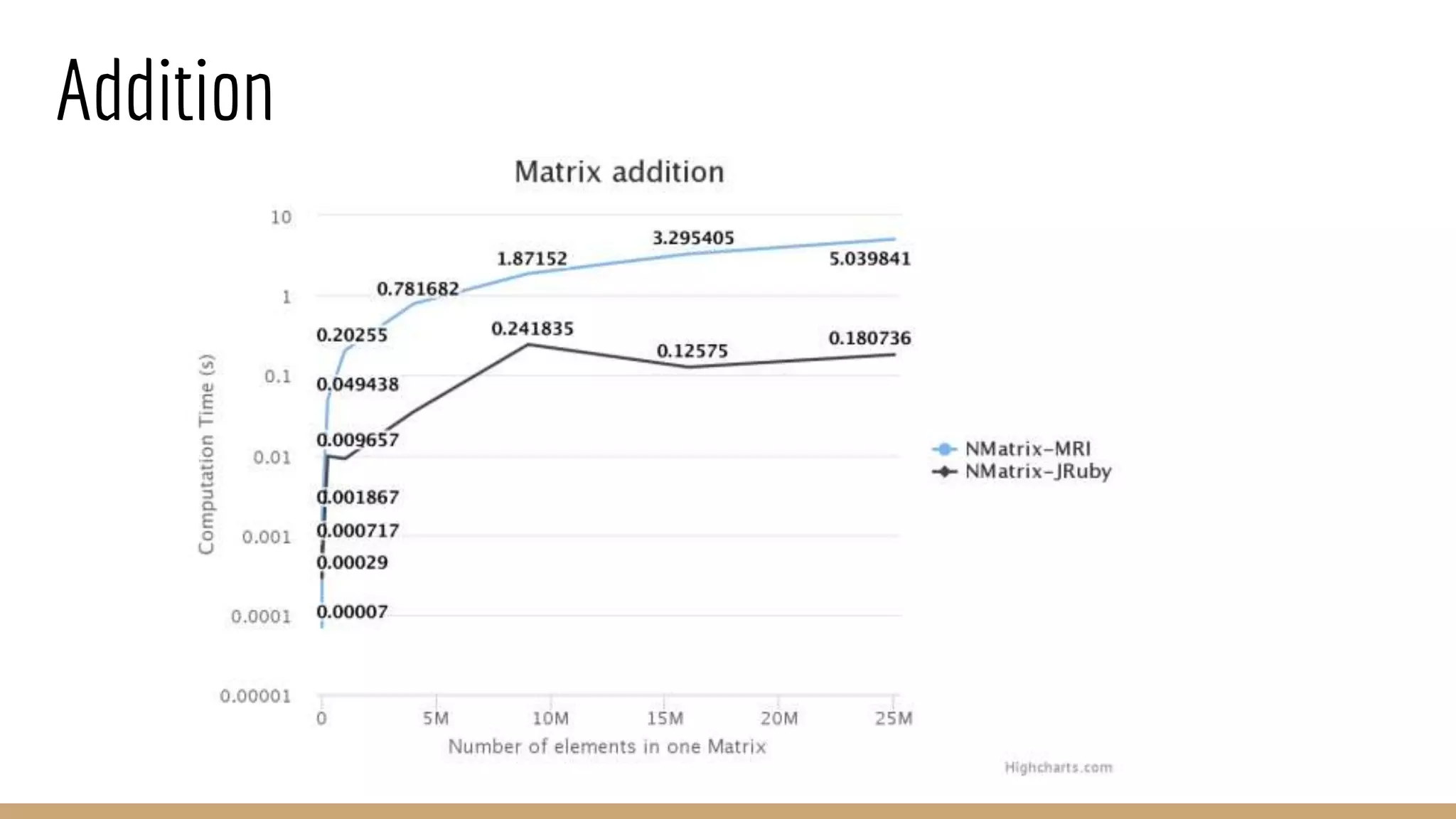
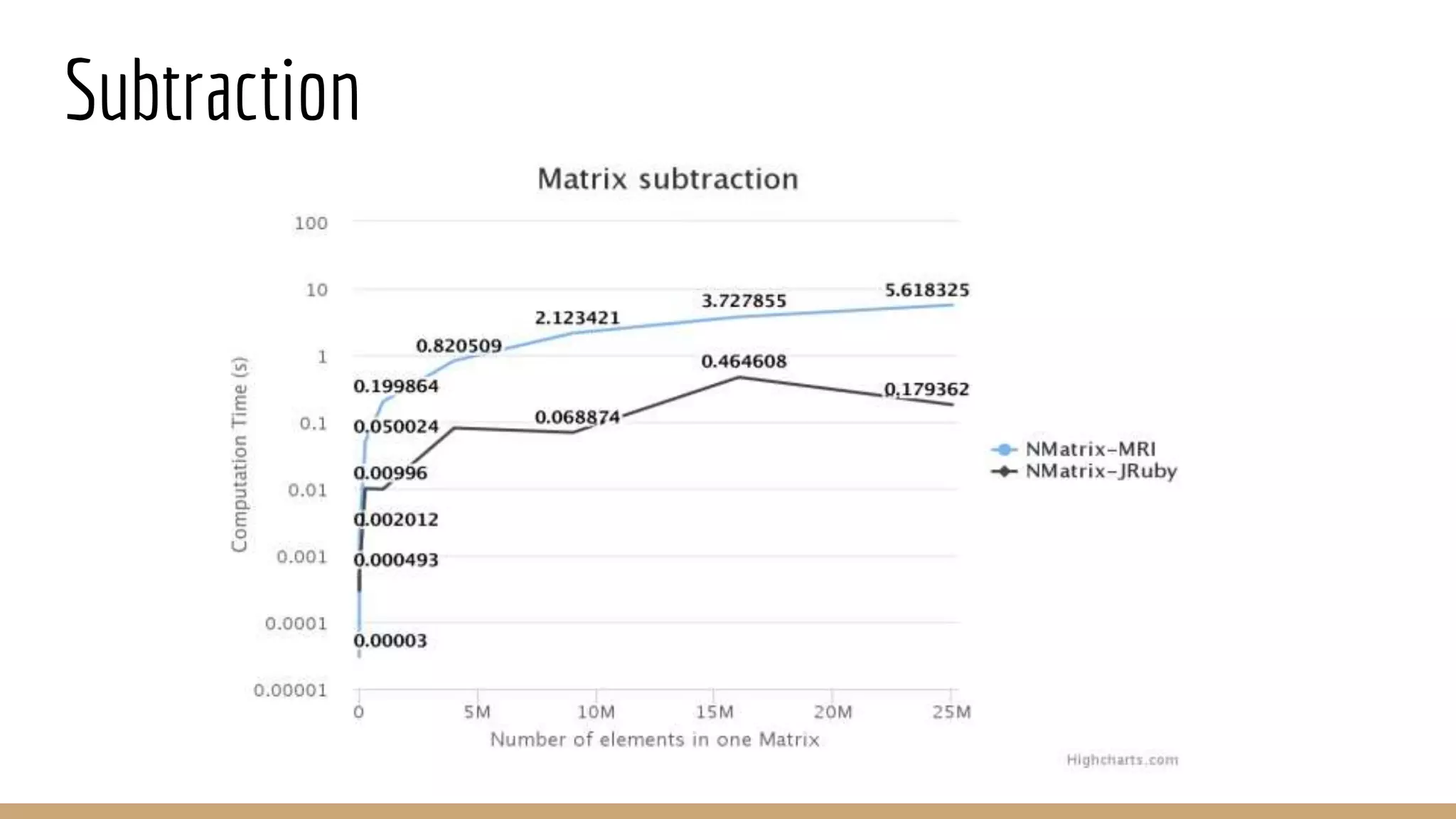
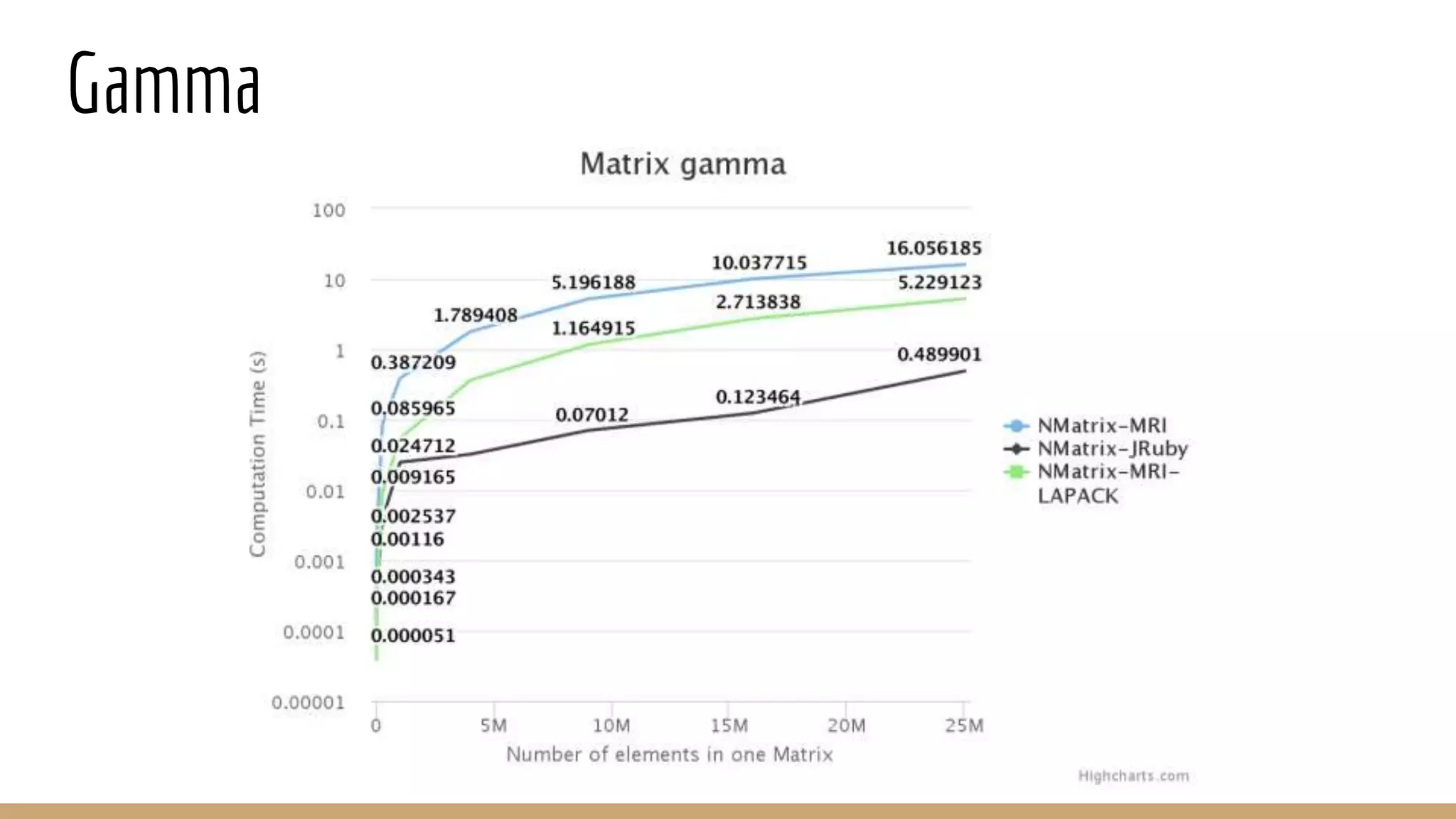
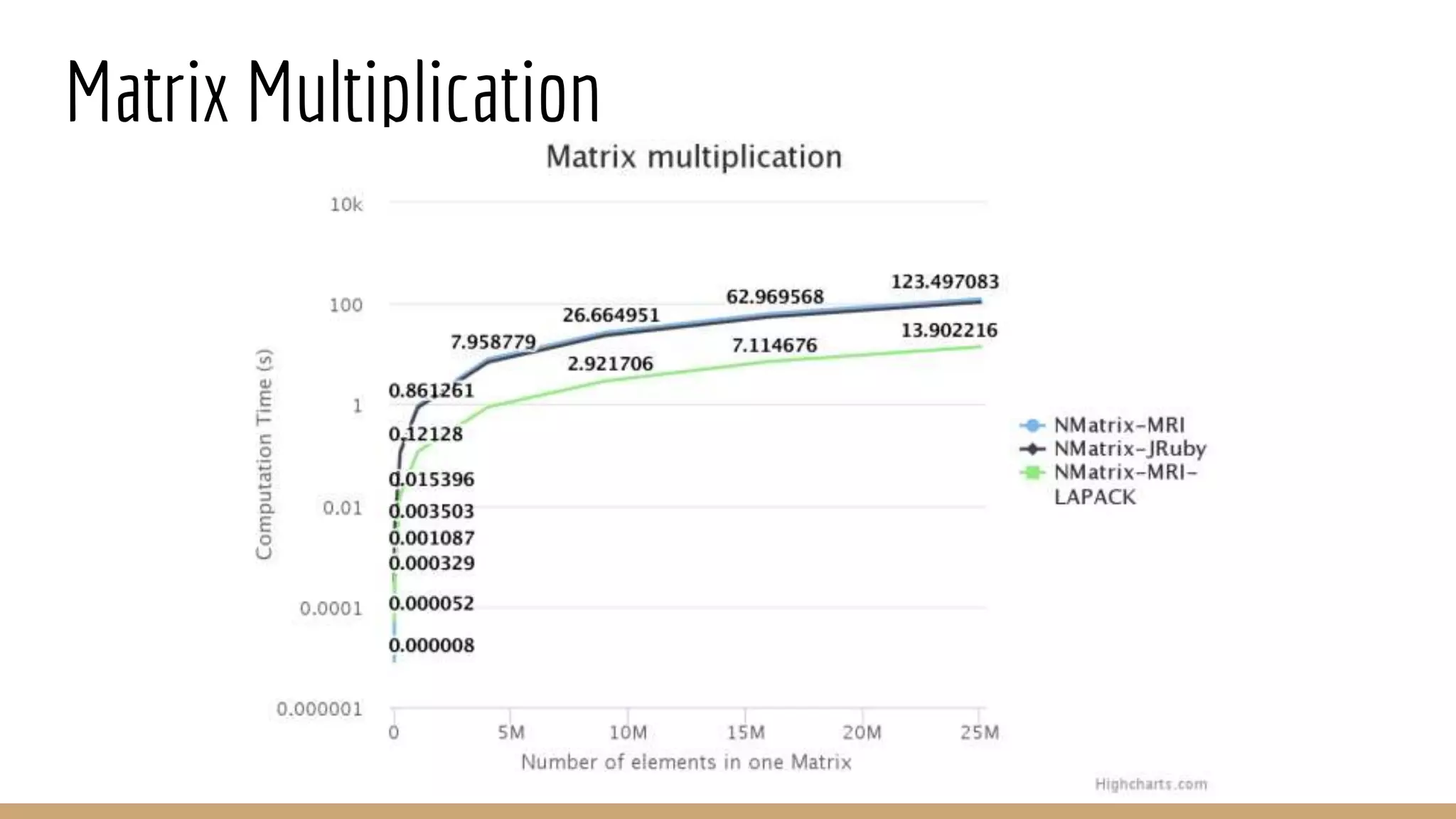
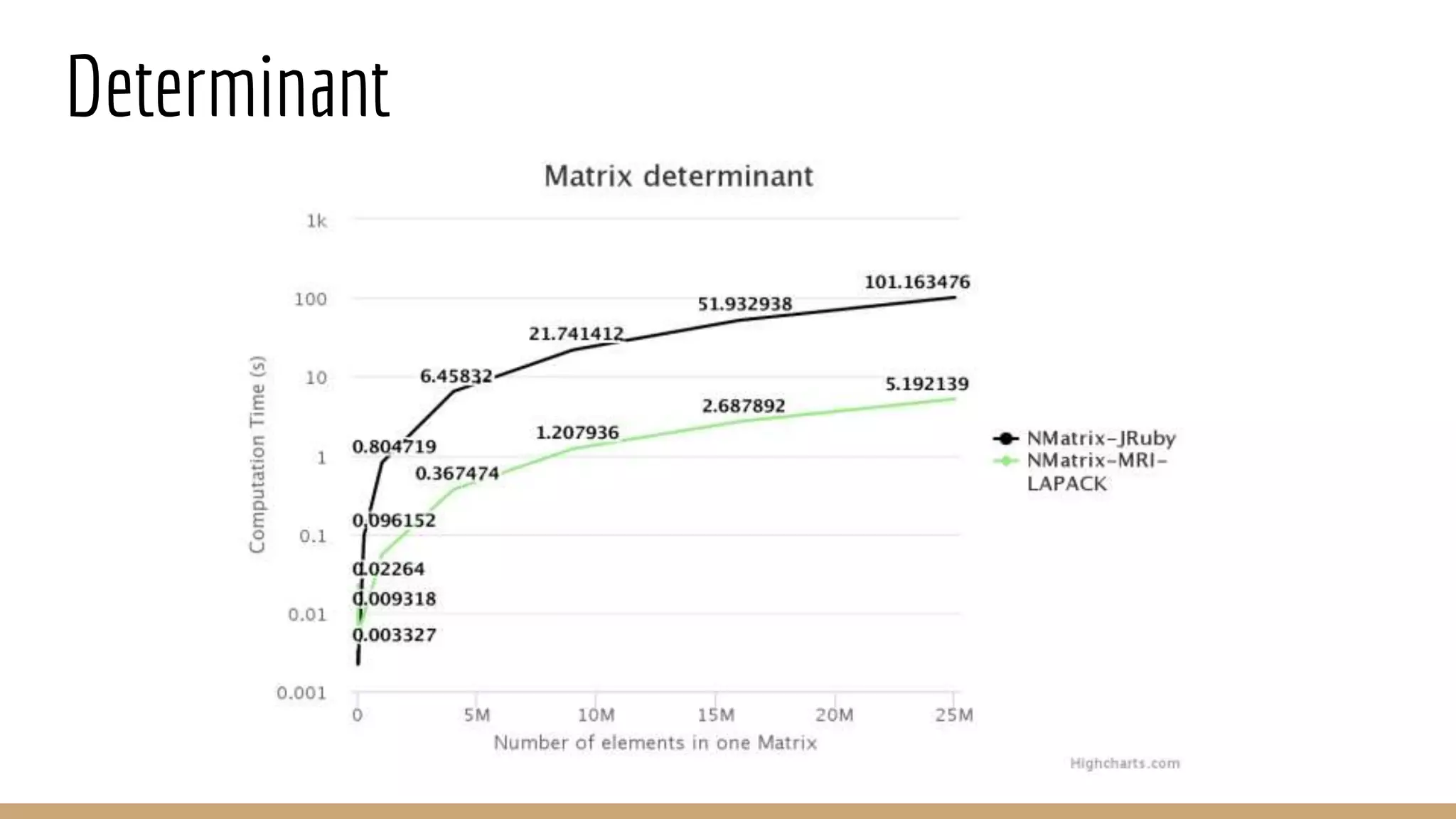
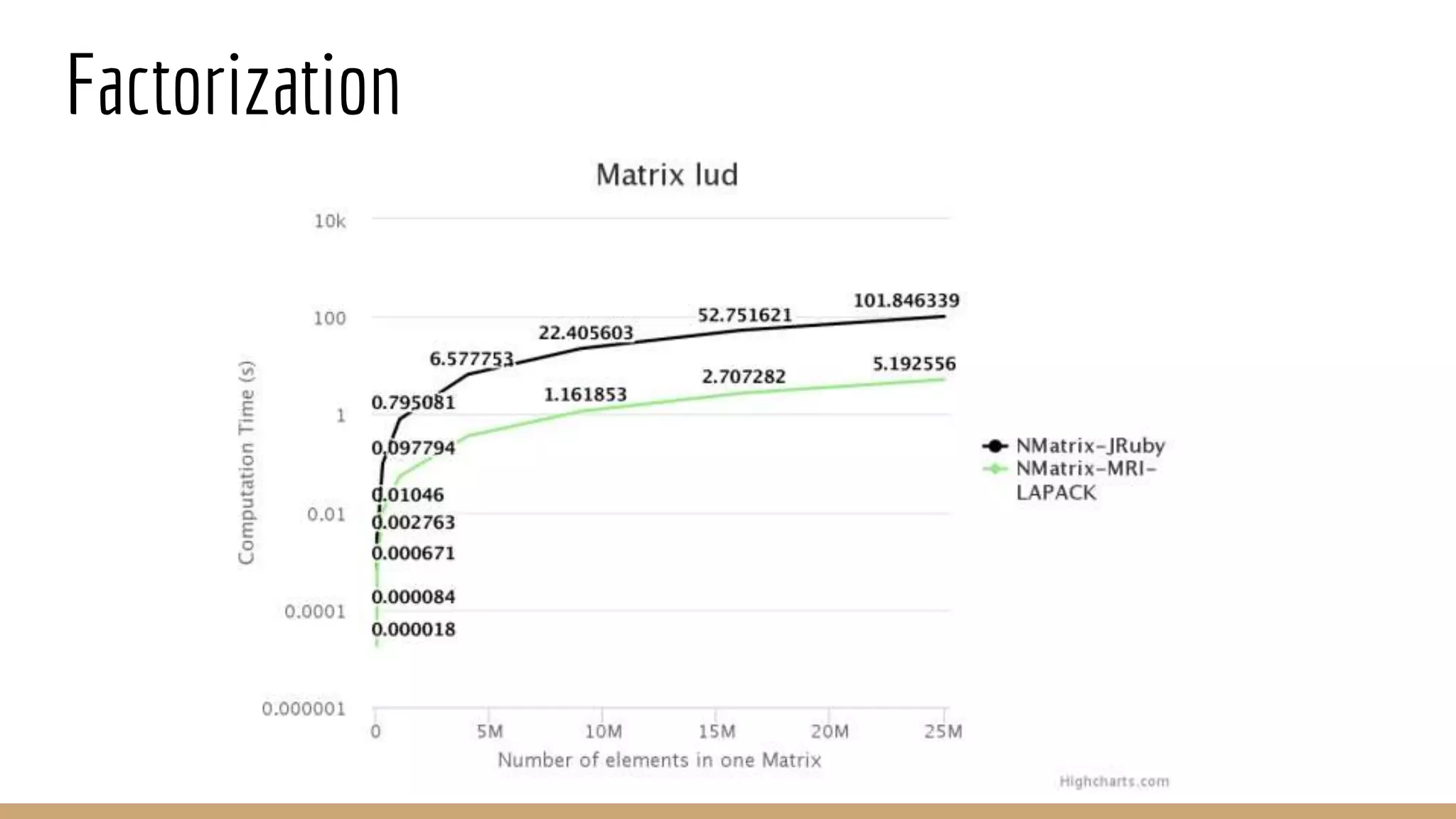
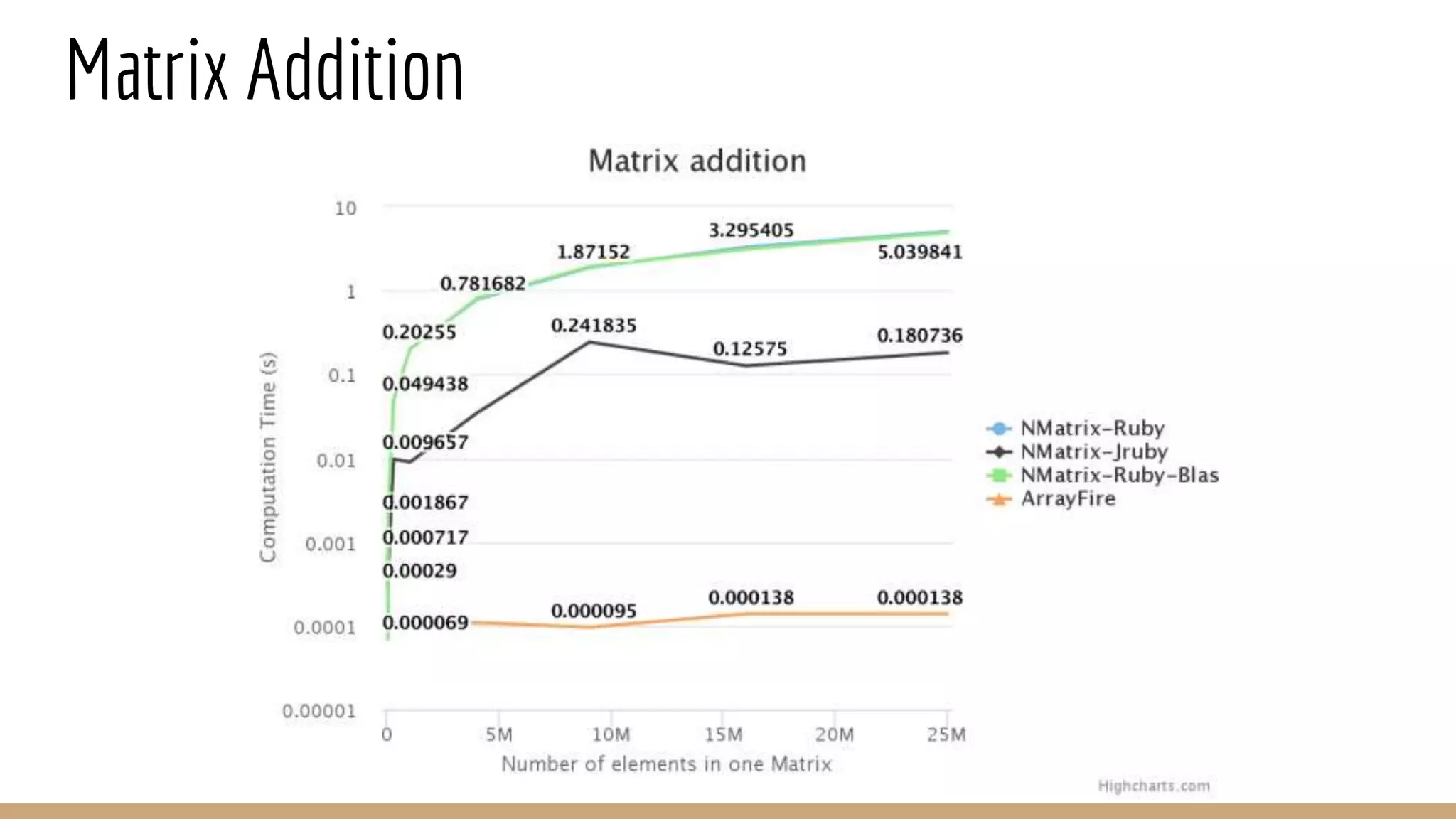
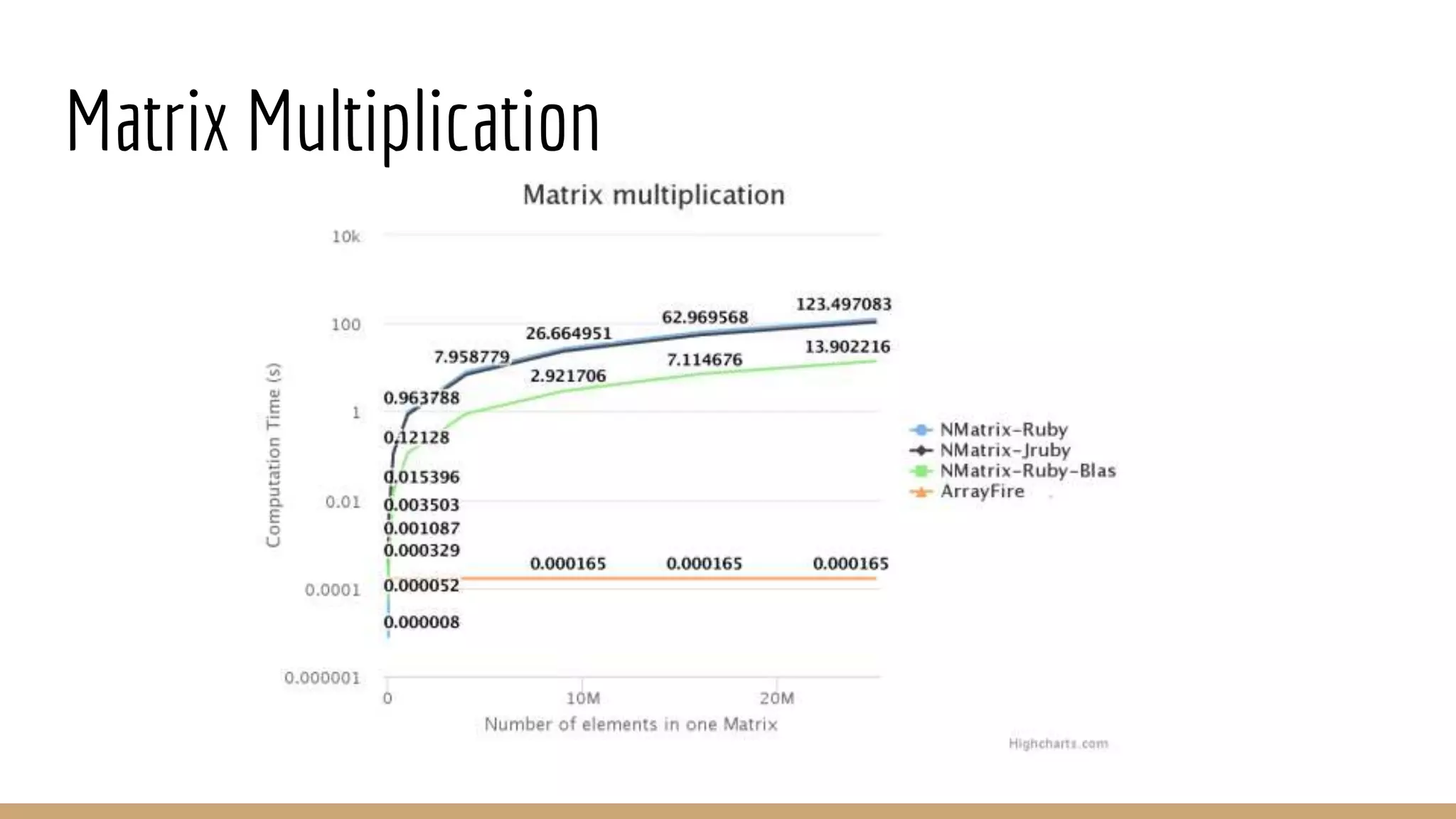
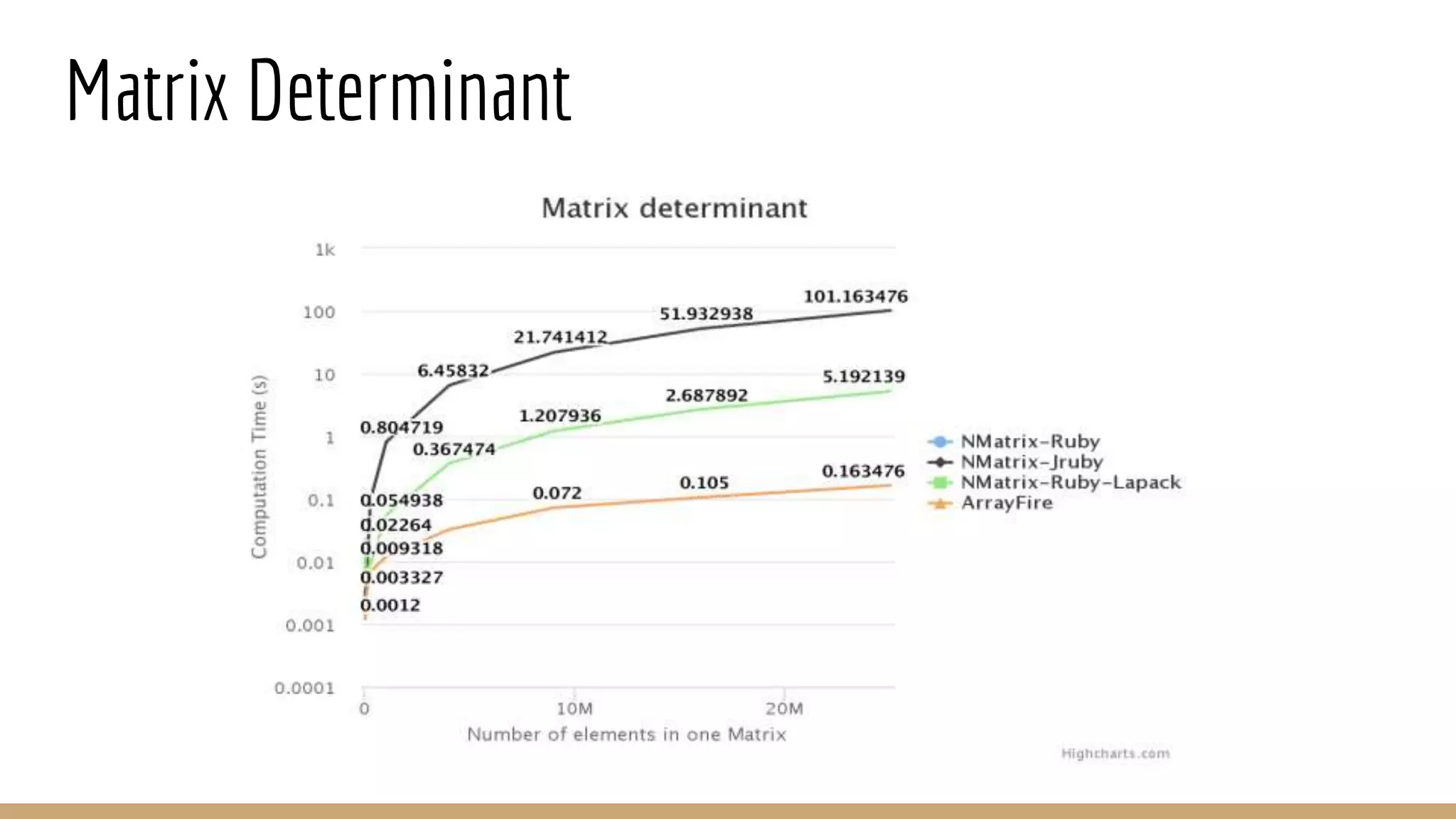
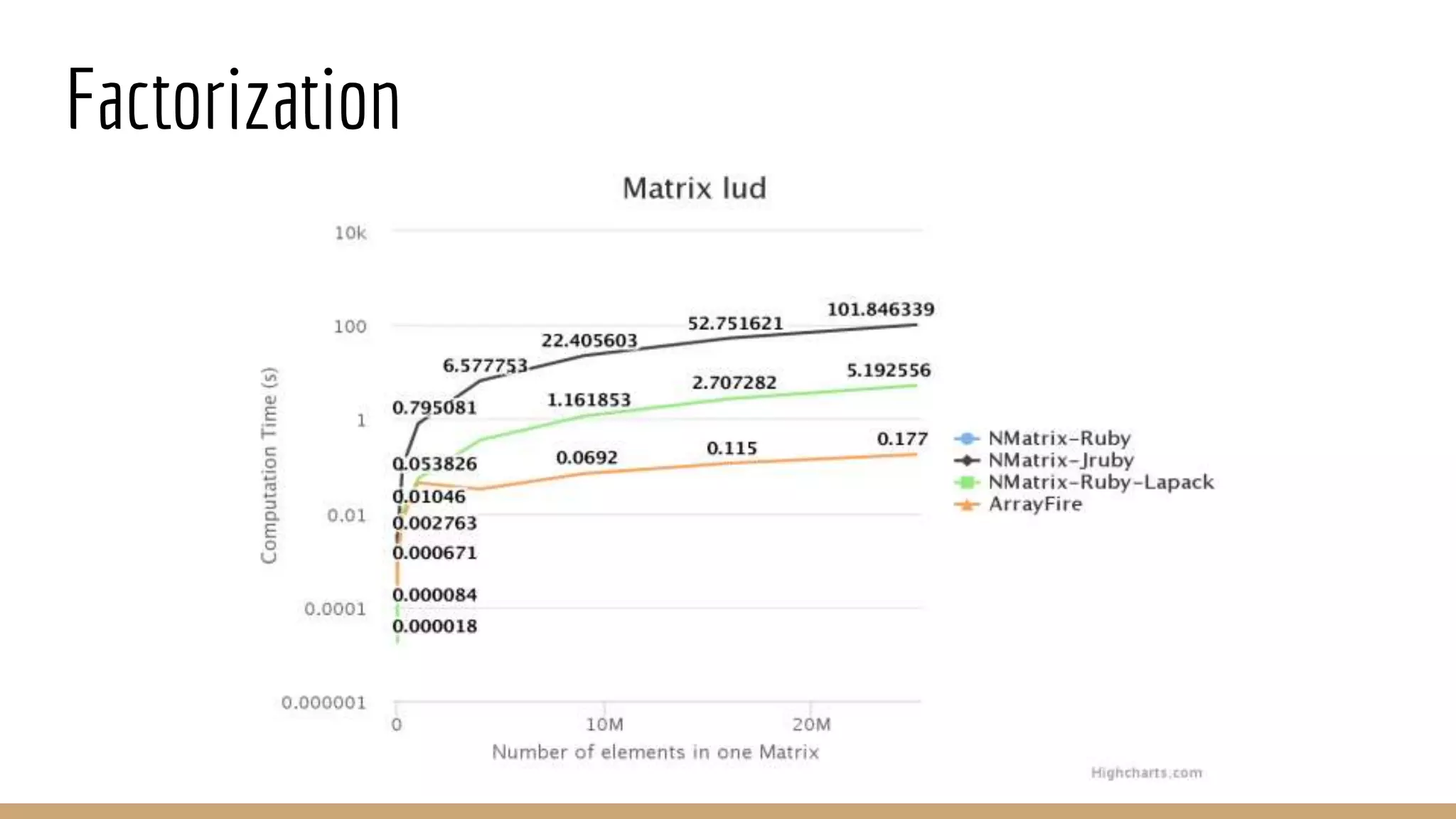
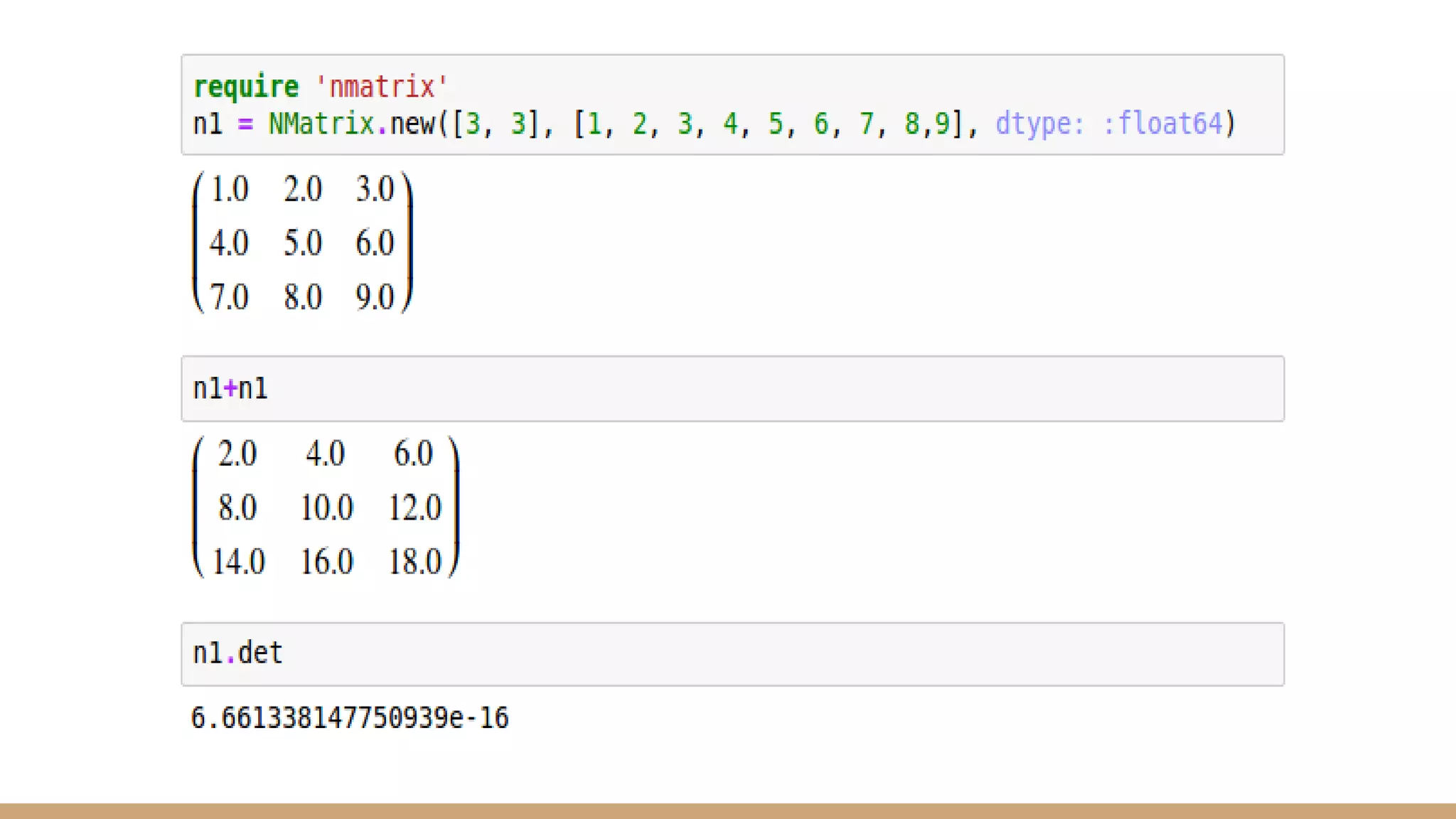
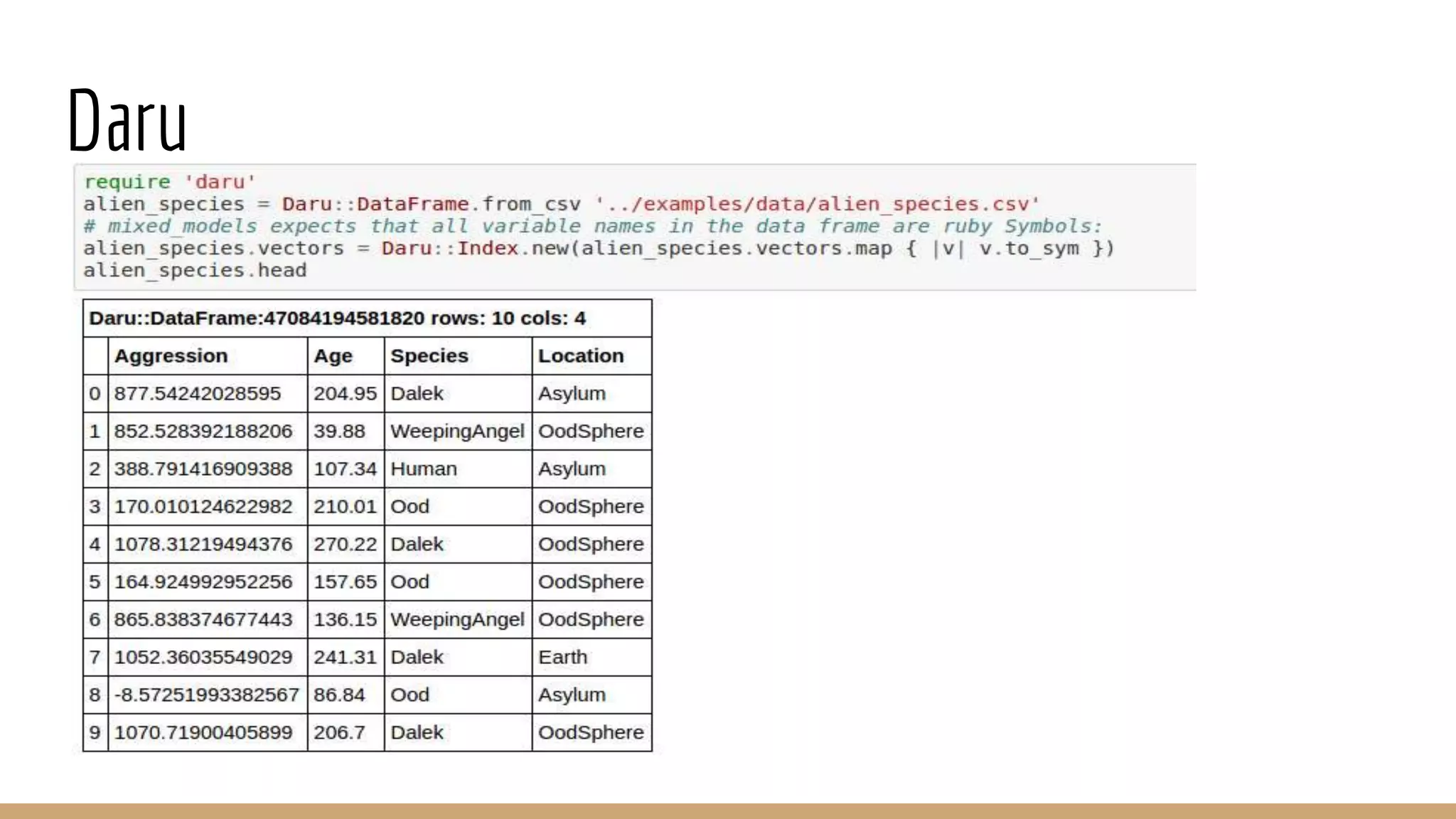
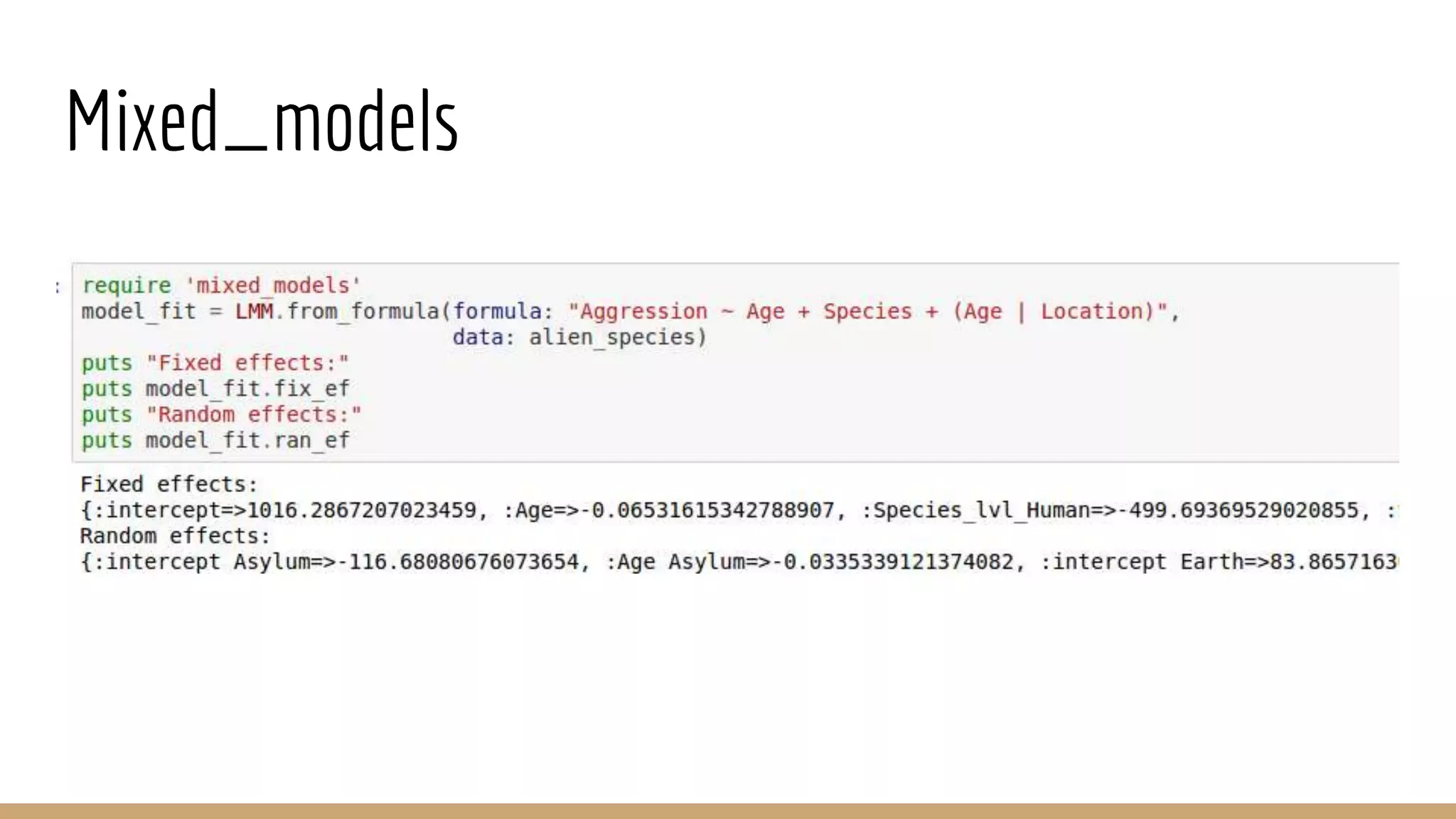
Scientific computing in Ruby can be improved using JRuby to leverage Java libraries and GPU processing. NMatrix, a numerical matrix library, was optimized for JRuby by reimplementing it to use efficient Java functions, improving speed by 1000x and reducing memory usage by 10x. Further optimizations for NMatrix-JRuby and integrating a general purpose GPU library called ArrayFire could enable faster matrix operations and transparent GPU processing for scientific applications in Ruby.









![Elementwise Operation ● Iterate through the elements ● Access the array; do the operation, return it ● [:add, :subtract, :sin, :gamma]](https://image.slidesharecdn.com/scientificcomputingonjruby-170129062115/75/Scientific-computing-on-jruby-19-2048.jpg)


![Autoboxing ● :float64 => double only ● Strict dtypes => creating data type in Java: not guessing ● Errors => that can’t be reproduced :P [ 0. 11, 0.05, 0.34, 0.14 ] + [ 0. 21,0.05, 0.14, 0.14 ] = [ 0, 0, 0, 0] ([ 0. 11, 0.05, 0.34, 0.14 ] + 5) + ([ 0. 21, 0.05, 0.14, 0.14 ] + 5) - 10 = [ 0.32, 0.1, 0.48, 0.28]](https://image.slidesharecdn.com/scientificcomputingonjruby-170129062115/75/Scientific-computing-on-jruby-23-2048.jpg)

![Ruby Code index =0 puts Benchmark.measure{ (0...15000).each do |i| (0...15000).each do |j| c[i][j] = b[i][j] index+=1 end end } #67.790000 0.070000 67.860000 ( 65.126546) #RAM consumed => 5.4GB b = Java::double[15_000,15_000].new c = Java::double[15_000,15_000].new index=0 puts Benchmark.measure{ (0...15000).each do |i| (0...15000).each do |j| b[i][j] = index index+=1 end end } #43.260000 3.250000 46.510000 ( 39.606356)](https://image.slidesharecdn.com/scientificcomputingonjruby-170129062115/75/Scientific-computing-on-jruby-26-2048.jpg)

![Java Code public class MatrixGenerator{ public static void test2(){ for (int index=0, i=0; i < row ; i++){ for (int j=0; j < col; j++){ c[i][j]= b[i][j]; index++; } } } puts Benchmark.measure{MatrixGenerator.test2} #0.034000 0.001000 00.034000 ( 00.03300) #RAM consumed => 300MB public class MatrixGenerator{ public static void test1(){ double[][] b = new double[15000][15000]; double[][] c = new double[15000][15000]; for (int index=0, i=0; i < row ; i++){ for (int j=0; j < col; j++){ b[i][j]= index; index++; } } } puts Benchmark.measure{MatrixGenerator.test1} #0.032000 0.001000 00.032000 ( 00.03100)](https://image.slidesharecdn.com/scientificcomputingonjruby-170129062115/75/Scientific-computing-on-jruby-28-2048.jpg)