Embed presentation
Downloaded 15 times
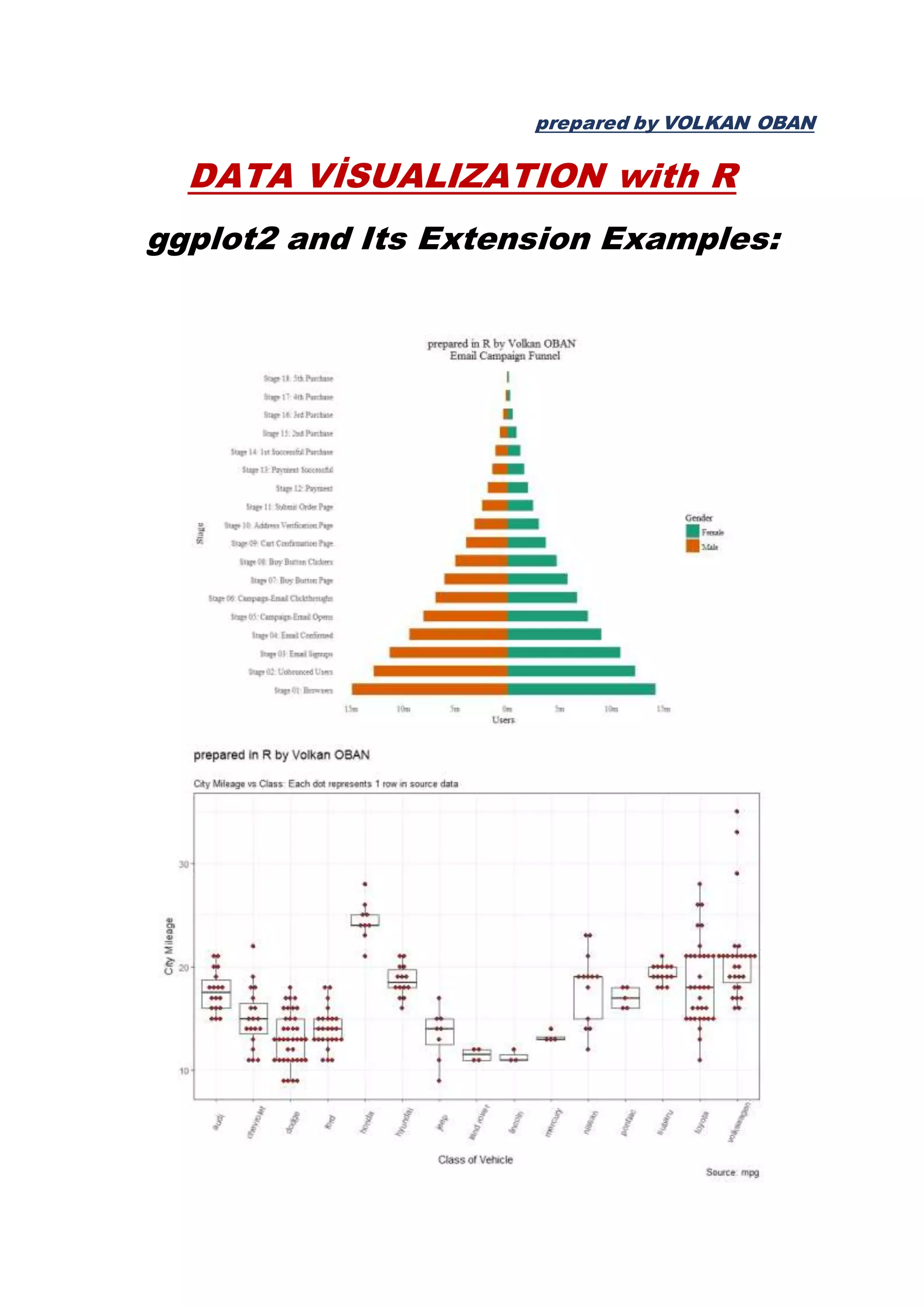
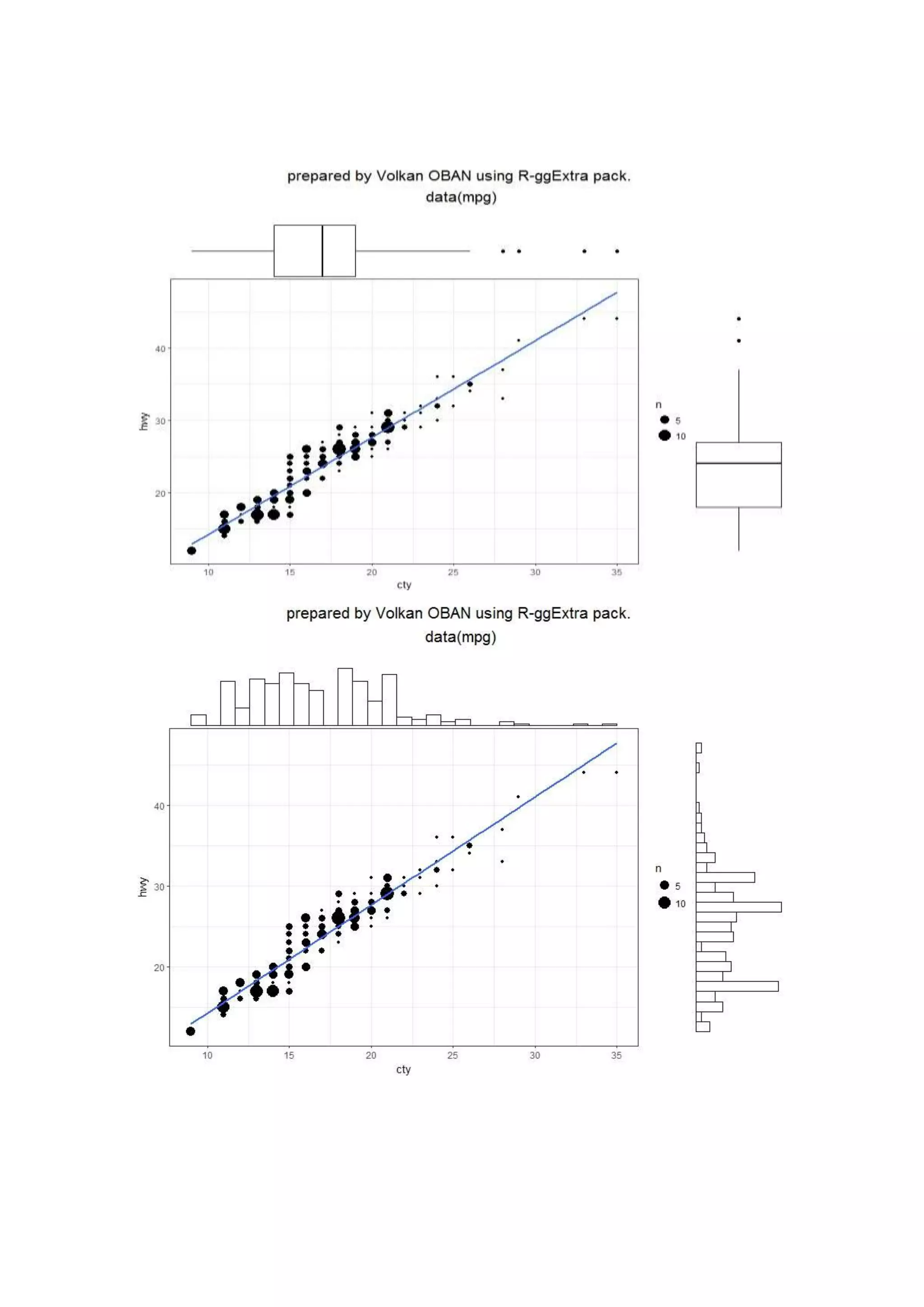
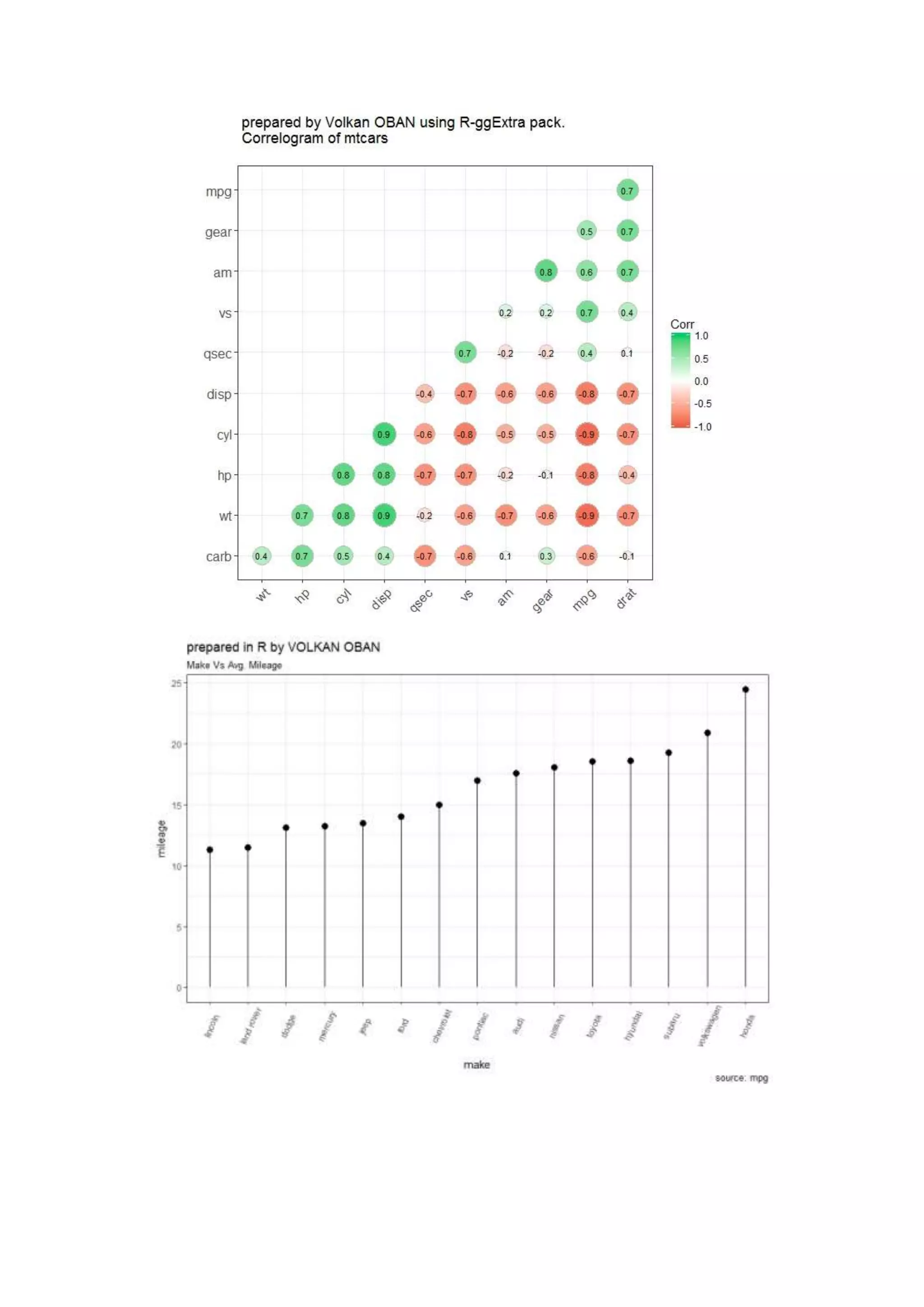
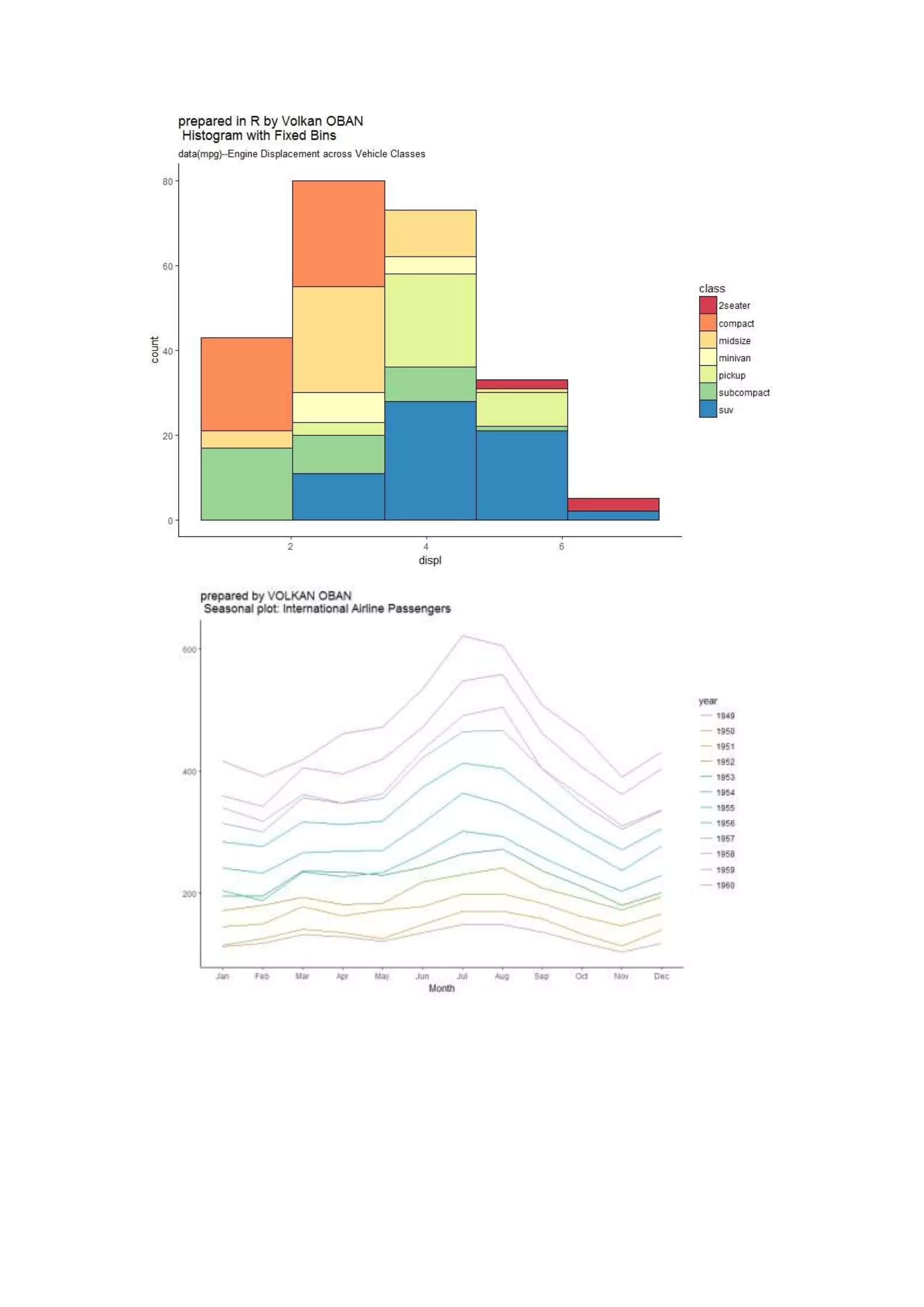
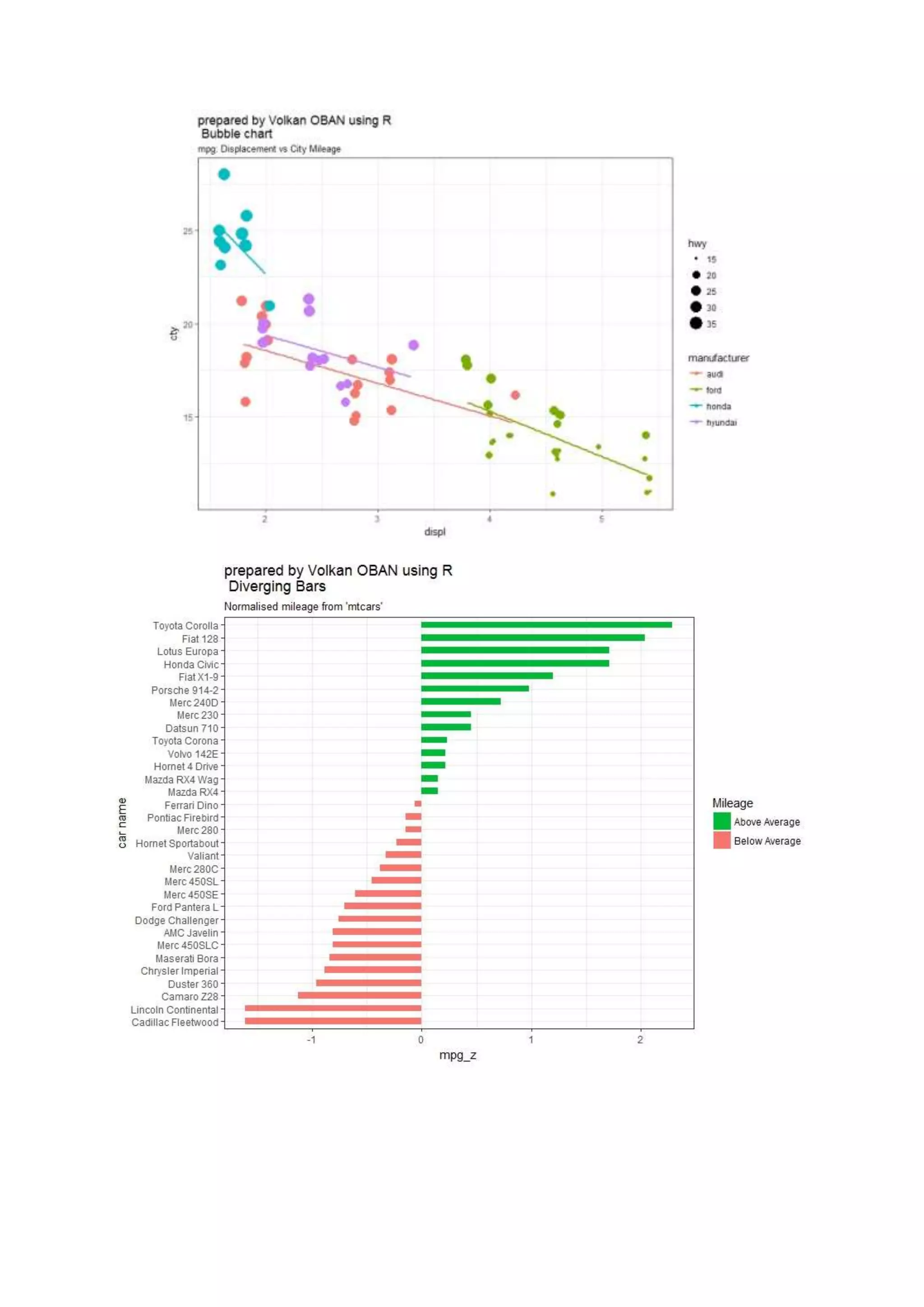
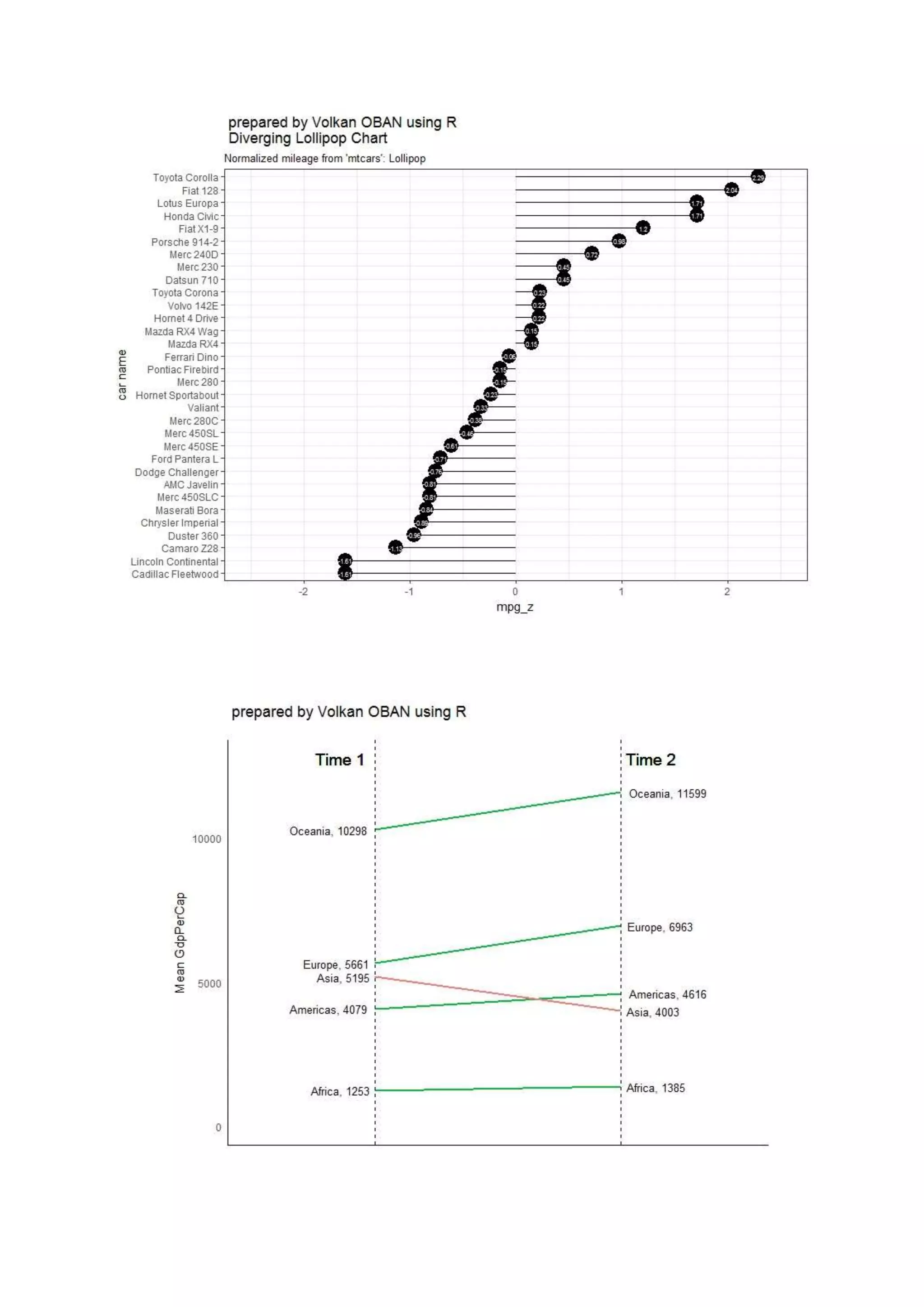
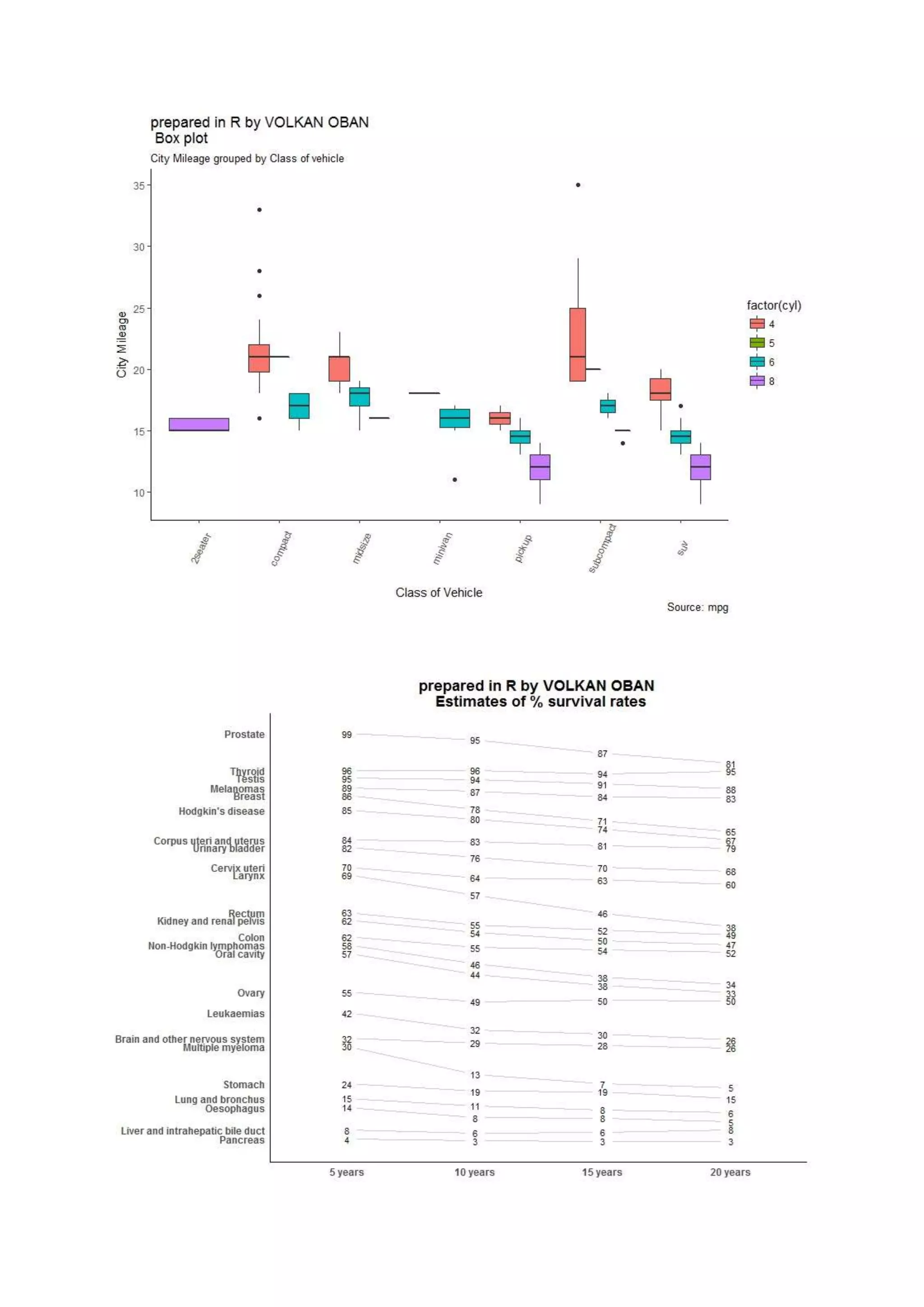
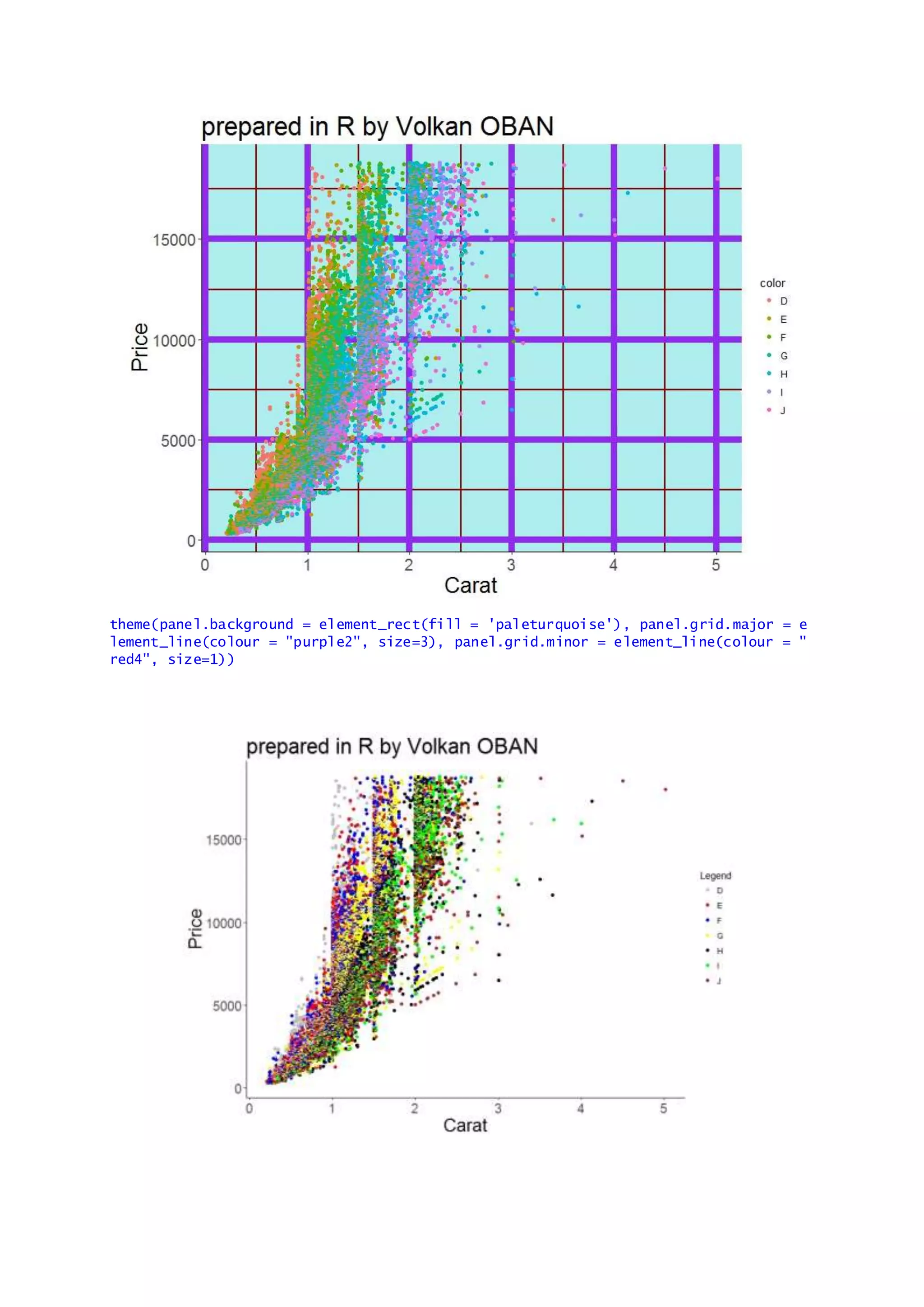
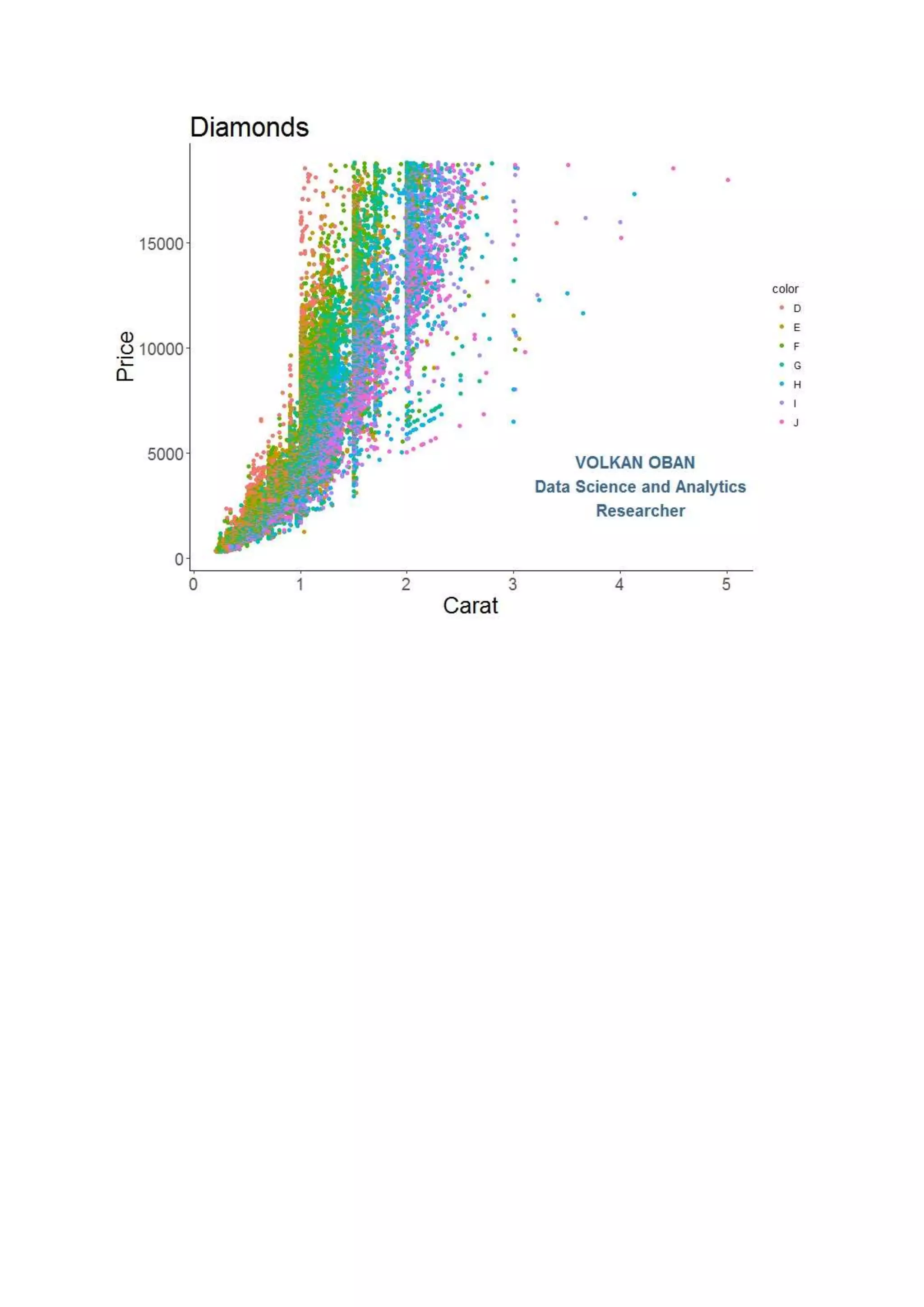
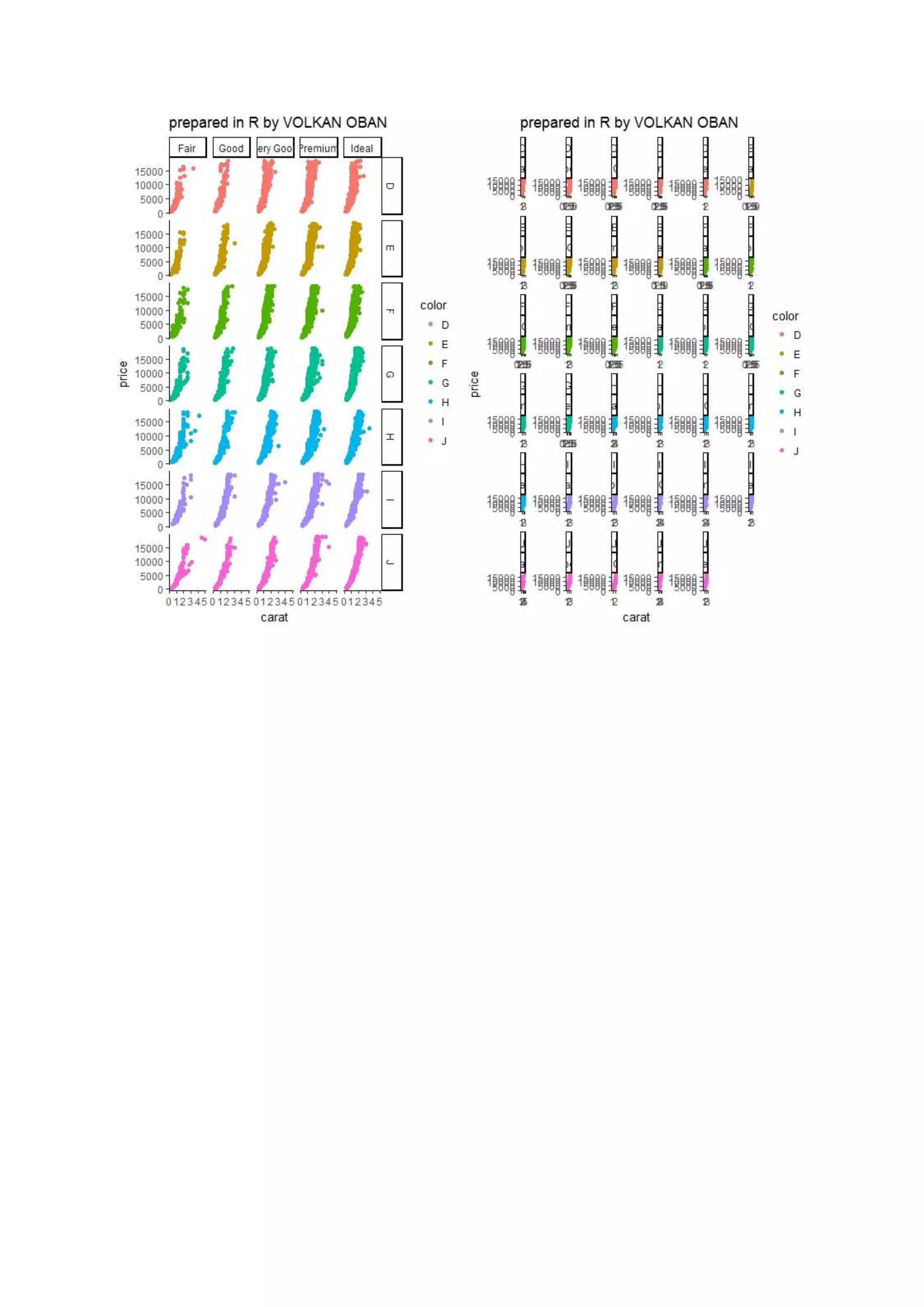
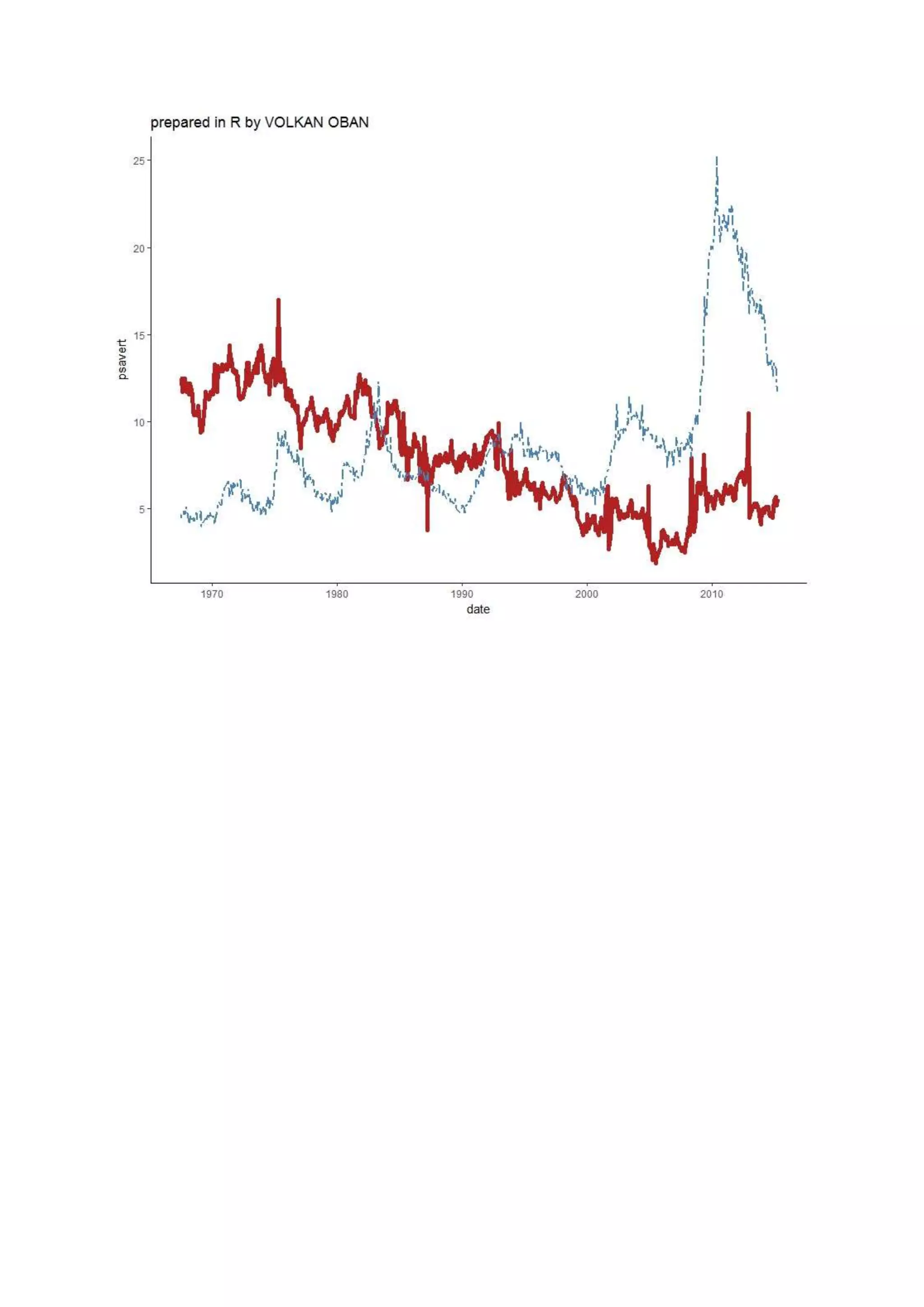
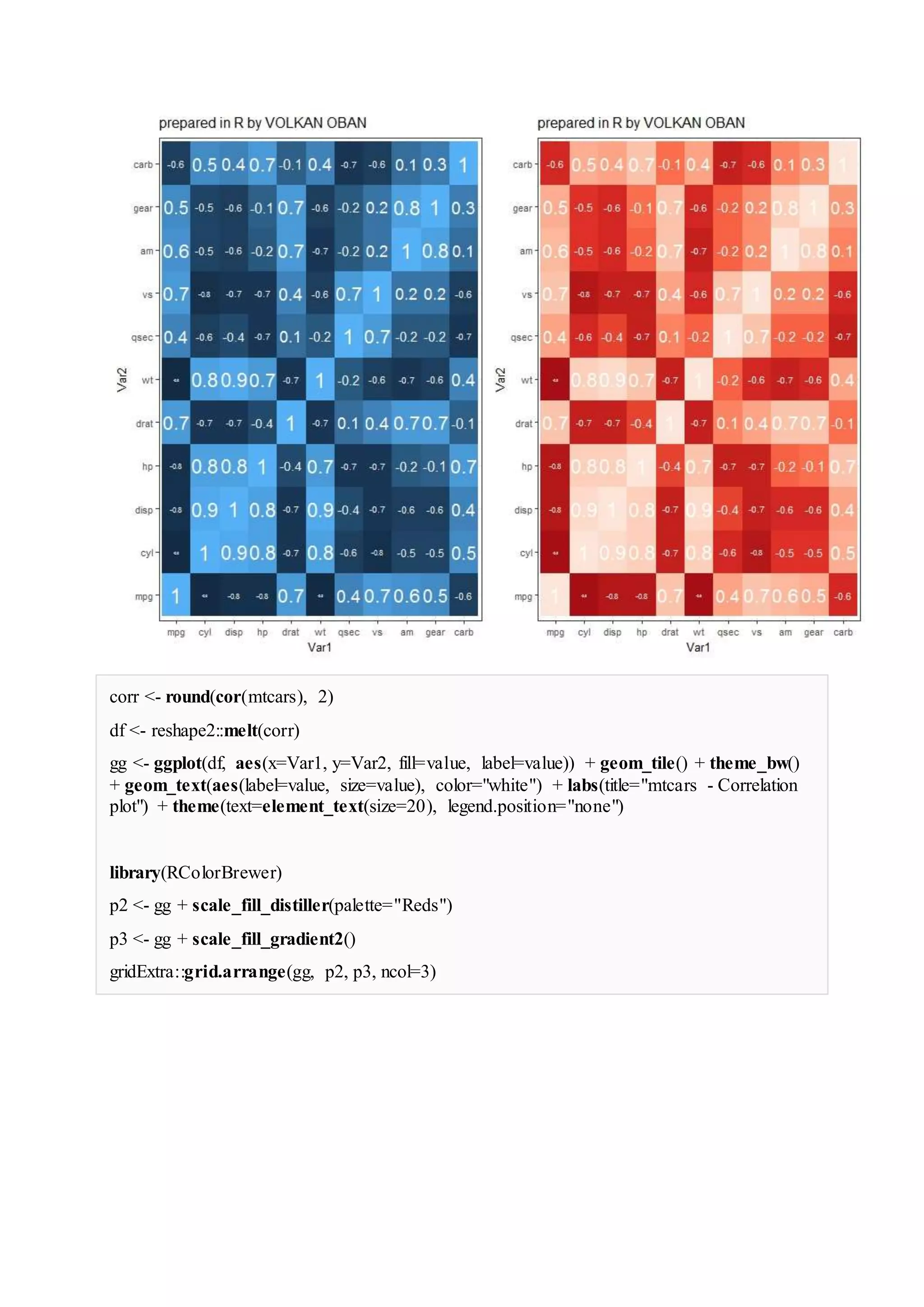
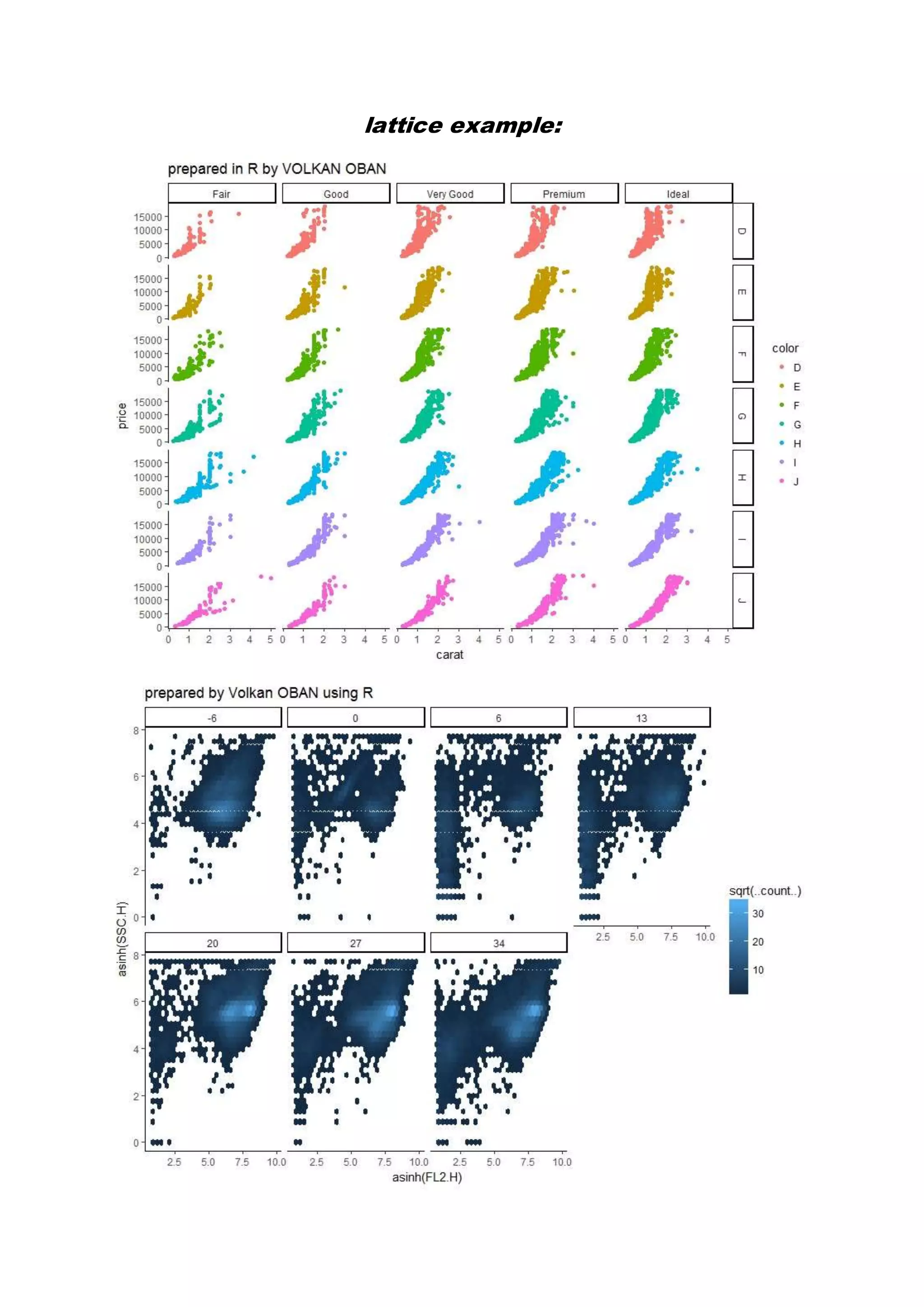
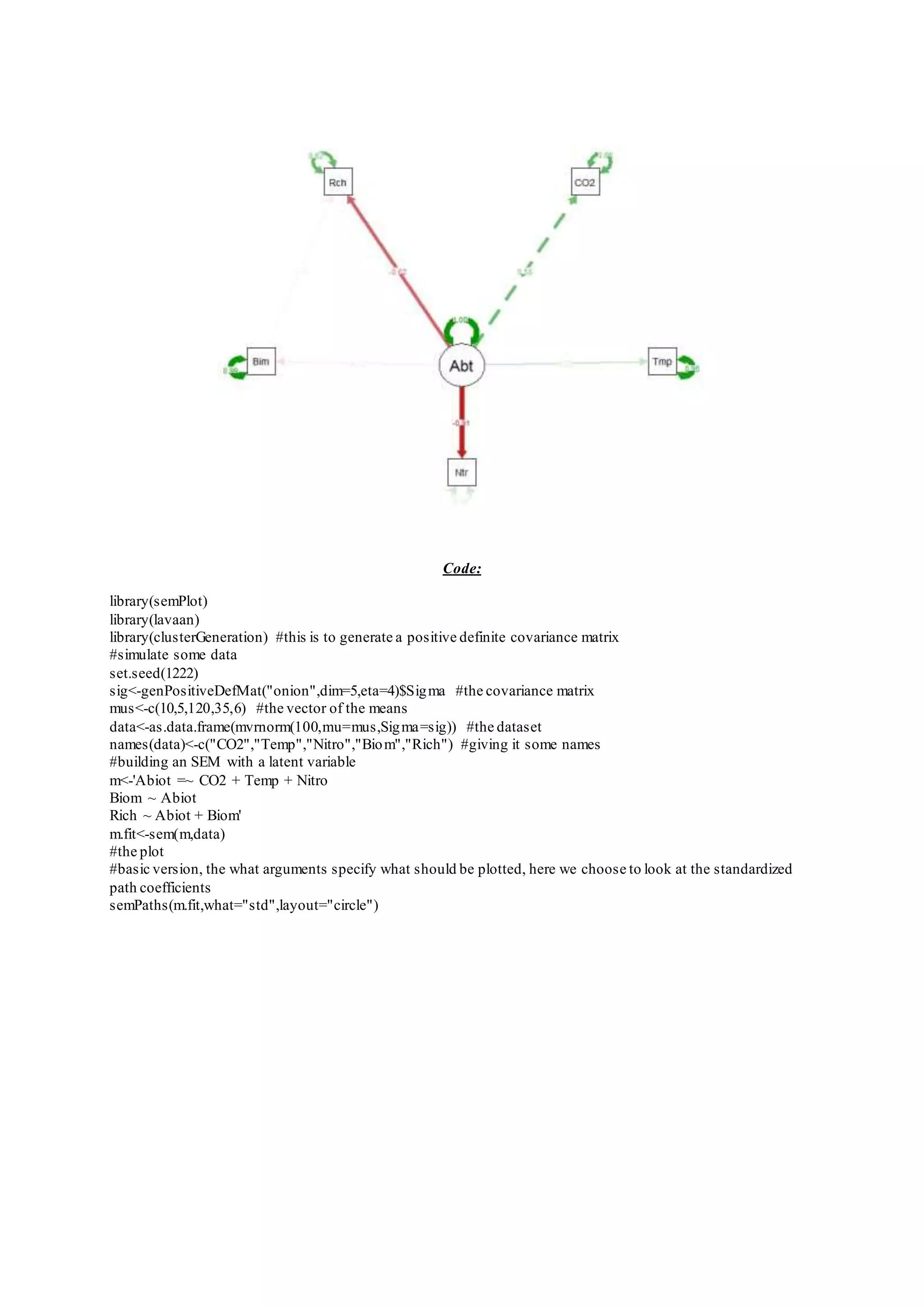


The document provides examples of data visualization in R using the ggplot2 library, including techniques for creating correlation plots and customizing themes. It also demonstrates how to simulate data and build a structural equation model (SEM) while showcasing the use of various libraries such as semplot and lavaan. Additionally, references to external resources for further learning are included.














