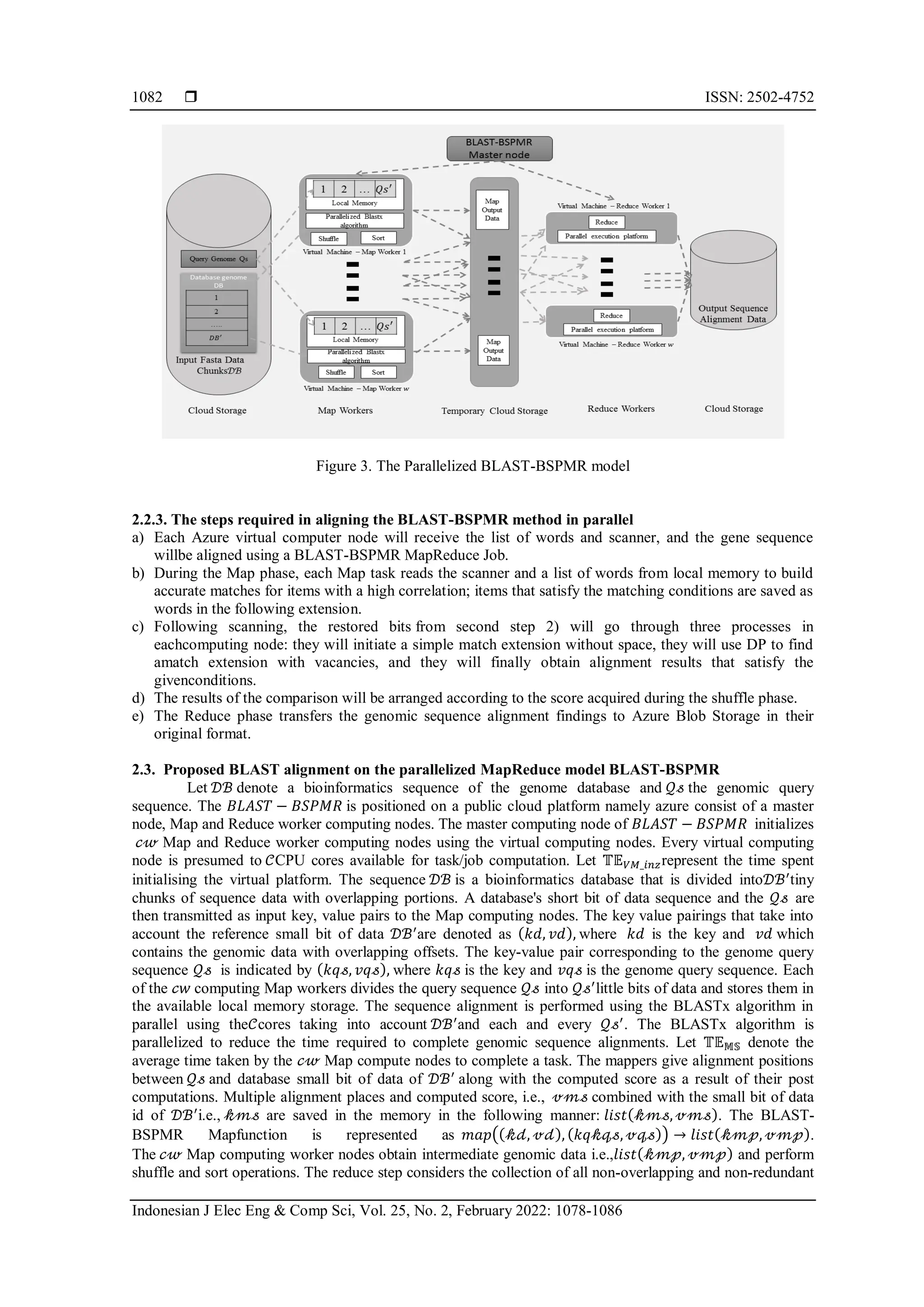
The document presents a novel parallel scheduling framework called blast-bspmr for bioinformatics applications, specifically focusing on genomic sequence alignment using the basic local alignment search tool (BLAST) algorithm. It details how this framework improves the existing Hadoop MapReduce architecture by allowing parallel execution of the map and reduce phases on the Microsoft Azure cloud platform, resulting in increased efficiency in processing large biological datasets. Experimental results demonstrate that the blast-bspmr framework significantly reduces computation time and enhances scalability compared to traditional methods.
![Indonesian Journal of Electrical Engineering and Computer Science Vol. 25, No. 2, February 2022, pp. 1078~1086 ISSN: 2502-4752, DOI: 10.11591/ijeecs.v25.i2.pp1078-1086 1078 Journal homepage: http://ijeecs.iaescore.com An efficient and robust parallel scheduler for bioinformatics applications in a public cloud: A bigdata approach Leena Ammanna1 , Jagadeeshgowda2 , Jagadeesh Pujari1 1 Department of Information Science and Engineering, SDM College of Engineering and Technology, Dharwad, India 2 Department of Computer Science and Engineering, Shri Krishna Institute of Technology, Bengaluru, India Article Info ABSTRACT Article history: Received Jul 7, 2021 Revised Dec 21, 2021 Accepted Dec 29, 2021 In bioinformatics, genomic sequence alignment is a simple method for handling and analysing data, and it is one of the most important applications in determining the structure and function of protein sequences and nucleic acids. The basic local alignment search tool (BLAST) algorithm, which is one of the most frequently used local sequence alignment algorithms, is covered in detail here. Currently, the NCBI's BLAST algorithm (stand- alone) is unable to handle biological data in the terabytes. To address this problem, a variety of schedulers have been proposed. Existing sequencing approaches are based on the Hadoop MapReduce (MR) framework, which enables a diverse set of applications and employs a serial execution strategy that takes a long time and consumes a lot of computing resources. The author, improves the BLAST algorithm based on the BLAST-BSPMR algorithm to achieve the BLAST algorithm. To address the issue with Hadoop's MapReduce framework, a customised MapReduce framework is developed on the Azure cloud platform. The experiment findings indicate that the suggested bulk synchronous parallel MapReduce-basic local alignment search tool (BSPMR-BLAST) algorithm matches bioinformatics genomic sequences more quickly than the existing Hadoop-BLAST method, and that the proposed customised scheduler is extremely stable and scalable. Keywords: Bioinformatics genomic sequence Hadoop Microsoft azure Scheduler parallelization This is an open access article under the CC BY-SA license. Corresponding Author: Jagadeeshgowda Department of Computer Science and Engineering, Shri Krishna Institute of Technology Bengaluru, India Email: hodcs.skitblr@gmail.com 1. INTRODUCTION According to research [1], [2], cloud computing has established itself as the future norm for data- intensive computing. According to [3], [4] cloud computing platforms allow on-demand access to shared, scalable, fault-tolerant, and reconfigurable computing resources with little administrative work and a low cost. In comparison to freestanding private computer clusters [5], [6] demonstrate how cloud computing platforms are a commonly desired and recognised method for running large data applications or high- performance computations (HPC). The cloud's resource management, virtual computing platforms, and elasticity all contribute to the ease with which data-intensive applications can be migrated to the cloud paradigm. Cai et al. [7], Okur and Büyükkeçeci [8] note that the frameworks required to enable the most efficient use of cloud resources at the lowest possible cost for data-intensive applications remain unresolved. Google’s MapReduce framework discussed in [9] is a commonly used approach for performing distributed computations on the cloud. White [10] and Hadoop [11] present that implementations of Hadoop are based on the MapReduce architecture and are capable of supporting large-scale data applications. The MapReduce model employs a two-stage execution mechanism. The initial stage entails splitting or dividing the data to be](https://image.slidesharecdn.com/15440-52181-1-pb-240902031046-71e8d34c/75/An-efficient-and-robust-parallel-scheduler-for-bioinformatics-applications-in-a-public-cloud-A-bigdata-approach-1-2048.jpg)
![Indonesian J Elec Eng & Comp Sci ISSN: 2502-4752 An efficient and robust parallel scheduler for bioinformatics … (Leena Ammanna) 1079 processed into little pieces. Each small bit of data is associated with a mapper. The mapper outputs ⟨𝐾𝑒𝑦|𝑉𝑎𝑙𝑢𝑒⟩ pairs that are arranged according to the 𝐾𝑒𝑦values. The Reduce workers are provided with the sorted values using the ⟨𝐾𝑒𝑦|𝑆𝑜𝑟𝑡𝑒𝑑𝐿𝑖𝑠𝑡(𝑉𝑎𝑙𝑢𝑒)⟩ method. The Hadoop distributed file system is used to store the results of the reduce workers hadoop distributed file system (HDFS). In most public cloud setups, the Map and Reduce workers are virtual machines (VMs). The researchers recognised the disadvantages of Hadoop MapReduce and examined several improvement strategies. Zhu et al. [12] discusses the use of Hadoop with CUDA to increase processing capacity and execution speed. Zhu et al. [12] proposes a research paradigm that facilitates appropriate use of graphics processing units (GPU). Dahiphale et al. [13] presents a cloud-based MapReduce model. Pipelining is used to improve execution performance and to enable elastic pricing. For scientific applications [14] proposed a framework based on parallel tempering called replica exchange statistical temperature molecular dynamics. Tang et al. [15] describe a unique dynamic Hadoop slot allocation technique for resolving the Hadoop resource provisioning problem that is not optimal. The result reported in Tang et al. [15] demonstrates that their technique significantly improves Hadoop's performance when handling many MapReduce jobs. The most often utilised technique is to increase execution effectiveness through scheduling practises, as described in [16]-[19]. Wang et al. [20] presented a method for computing "Imprecise Applications" using MR frameworks. Reduce is run after the Map step in predictable MapReduce applications. In the case of inaccurate applications, the Reduce stage might be initiated using the Map stage's partial findings. Applications such as word frequency statistics and hot-word identification are considered imprecise. Executing inaccurate apps on well- known machine learning frameworks like Hadoop introduces latency. Running erroneous apps might result in additional expenditures on public cloud systems, where users are charged for all computations and storage services consumed. To address this shortcoming incorporated the check phase into the MapReduce framework in order to reduce costs and execution latencies [20]. In MR frameworks, iterative applications and certain graph-based applications performed poorly. Google introduced the Pregel framework for cloud computations in [21] for similar applications. Pregel is built on valiant's bulk synchronous parallel (BSP) computation model [22]. Kajdanowicz et al. [23] demonstrate that the Pregel framework outperforms MapReduce when calculating graph-based applications. Apart from the serial execution mechanism utilised in MR, several concerns remain unresolved, including node failure management capabilities, scheduling strategies, and multiway join processes. The authors provide a framework for parallel computation MapReduce (PMR) in public cloud environments in this article. The MapReduce architecture of the BLAST-BSPMR enables parallel computation to accelerate the execution of the BSP model. The BLAST-BSPMR implements Map and Reduce workers by utilising Microsoft Azure VM computing environments. Multicore processors enable parallel computation in these virtualized computing environments. The BLAST-BSPMR proposal leverages this parallel execution feature to drastically lower the Map and Reduce worker nodes' computation times. In comparison to other MapReduce frameworks like Hadoop, the BLAST-Reduce BSPMR's phase is initiated when two or more worker nodes have completed their jobs. The BLAST-BSPMR function shown here is used to calculate the results of bioinformatics BLASTx applications. Several strategies and scheduler are proposed by researchers to improve the performance of the MapReduce cloud computing framework proposed by Google in Dean et al. [9]. The work presented in Zhu, et al. [12] endures the closest similarity to our work presented here. Using CUDA codes the Map and Reduce worker tasks execute parallely on the GPU’s is achieved. The integration of Hadoop and GPU is achieved using the Hadoop Streaming, Pipes, JCuda and JNI approaches. The experimental investigation given in Jie Zhu et al. [12] demonstrates the effectiveness of the suggested approach on a private heterogeneous cloud environment using the word count application. The execution efficiency of the JCuda approach over Hadoop Streaming, Pipes and JNI is also proved. The major drawback of this approach is that such computational models are not suited for public cloud environments as GPU based VM environments aregenerally not offered. Public cloud environments like Amazon EC2 that offer such GPU based virtualized computing environments provide it at very high costs. Dahiphale et al. [13] described the drawbacks of the conventional MapReduce frameworks as followsthe MapReduce framework adopts a sequential processing of the Map and Reduce stages. The scalability of the MapReduce is limited. The MapReduce framework provides no support for flexible pricing options. The MapReduce model provides no support for computing streaming data. To overcome these drawbacks a pipelined model is presented to parallelize the execution of the Map and Reduce phase. The MapReduce model proposed in [13] is realized on the Amazon public cloud. The spot instance offering of the Amazon cloud allows flexible pricing. The experimental study given here demonstrates the efficiency of the pipelining-based MapReduce model when compared to the conventional MapReduce model using the word count application. The major drawback of the model proposed in [13] is that the locality optimization is not](https://image.slidesharecdn.com/15440-52181-1-pb-240902031046-71e8d34c/75/An-efficient-and-robust-parallel-scheduler-for-bioinformatics-applications-in-a-public-cloud-A-bigdata-approach-2-2048.jpg)
![ ISSN: 2502-4752 Indonesian J Elec Eng & Comp Sci, Vol. 25, No. 2, February 2022: 1078-1086 1080 considered and hosting of additional data dependent applications like Smith Waterman and other bioinformatics application cannot be executed in a pipelined fashion. The computation of certain data intensive applications like graph applications and iterative applications on MapReduce frameworks exhibit high computation time and cost. To support such applications, Google presented a proprietary cloud computing framework named Pregel that embraces the 𝐵𝑆𝑃 computing model [21]. In Pregel, the graph computations are achieved using a set of super-steps. A super step is used to execute the user defined application or function in a parallel fashion using the data item from the database. Each data item from the database behaves as an agent. The Pregel system adopts vertex-centric execution strategy. The computation of each data item has a graph like representation in 𝐵𝑆𝑃. The vertexes in the Pregel deactivate post the computation operation and are reactivated only if additional data items are presented to them. Once all the vertices are deactivated the computation is said to be complete. The local storage of the data items in the nodes executing the computation poses a problem. In the case the data item is large then a spilling- to-disk technique needs to be in [23]. The MapReduce framework is been adopted by the Hadoop framework in Nguyen et al. [24] for computing on the cloud platform. The MapReduce paradigm employs a two-phase technique. The first phase divides the input data into little bits of data to be processed. Each small bit of data is associated with a mapper. The mapper outputs a<Key, Value> pair that is sorted according to the Key values. Thereducer takes these sorted values as the <Key│SortedList (Value)>. These sorted values are stored in the Hadoop Distributed File System. Schatz [25] and CloudBurst make use of the Hadoop MapReduce framework as the computing platform. These alignment tools perform effectively for small base pair alignments requiring single-gap or un- gapped alignment. However, when huge base pairs are considered, these aligners perform poorly. Because all present cloud framework sequence aligners take the Hadoop framework into account, Hadoop suffers when iterative applications are hosted on the cloud framework. When considering multiway joins, Hadoop performs poorly. Since Hadoop framework executes sequentially, again there is degradation in the performance. The 𝐵𝑆𝑃 based Pregel framework exhibits lower computation time for selective applications. In this paper the 𝐵𝐿𝐴𝑆𝑇 − 𝐵𝑆𝑃𝑀𝑅 framework incorporates the MapReduce architecture and an execution strategy is conducted in parallel fashion as observed in the 𝐵𝑆𝑃. 2. METHOD A cloud platform, BLAST-BSPMR is proposed that allows to apply bioinformatics application like gene sequencing. The BLAST-BSPMR uses MapReduce framework which works on a cloud computing environment. Azure VM is the Azure infrastructure as service (IaaS) which is used to deploy persistent VMs. Thus, the Map and Reduce worker nodes in BLAST-BSPMR are deployed on Azure VMs. Alignment of genomic sequences is performed using the BLASTxmethod. Here in BLASTx, a DNA query is compared to a protein database. The BLAST-BSPMR algorithm performs sequence alignment in two stages, namely the Map and Reduce phases. SinceHadoop framework is sequential, only when the Map phase is completed the Reduce phase gets executed. To prevent sequential execution, BLAST-BSPMR considers parallelizing the Map and Reduce phases. The Map and Reduce functions are meant to be executed in parallel and to make optimal use of the cores available in the worker VMs. 2.1. Basic local alignment search tool (BLAST) algorithm Basic local alignment search tool (BLAST) is used to compare a query sequence with the database sequence to find similar sequences in the database. The BLAST consists of various sequence searching programmes (including BLASTn, tBLASTn, BLASTx, BLASTp, tBLASTp, and tBLASTx). Each application varies in its functionalities, but the core of the algorithm for all of them is basically the same. The BLAST algorithm consists of three steps. In the initial step, called as building the word list, the query sequence is split intosmall words of fixed size (w-mers), often w being 11. Therefore, if the length of the query sequence is n, then list of n-w+1 words are being constructed. This step is illustrated in Figure 1. The second stage, referred to as the scan for hits, is where the BLAST algorithm looks for matches to the words (referred to as hits) in the database sequence. The BLAST algorithm employs the 'two-hit' strategy. If two non-overlapping word pairs are found to be within a distance D of one another, an extension is initiated. The next stage, dubbed extend the hits, involves the BLAST algorithm expanding the results in order to find longer related segment pairs. When the "two-hit" requirement is met, a gap-free extension in two directions is performed to locate an alignment known as a high-scoring segment pair (HSP). Figure 2 gives an example of gap-free extension. This is the seed-and-extend heuristic strategy for identifying high-scoring gene sequence alignments between the genomic query sequence and the database's genomic sequences.](https://image.slidesharecdn.com/15440-52181-1-pb-240902031046-71e8d34c/75/An-efficient-and-robust-parallel-scheduler-for-bioinformatics-applications-in-a-public-cloud-A-bigdata-approach-3-2048.jpg)


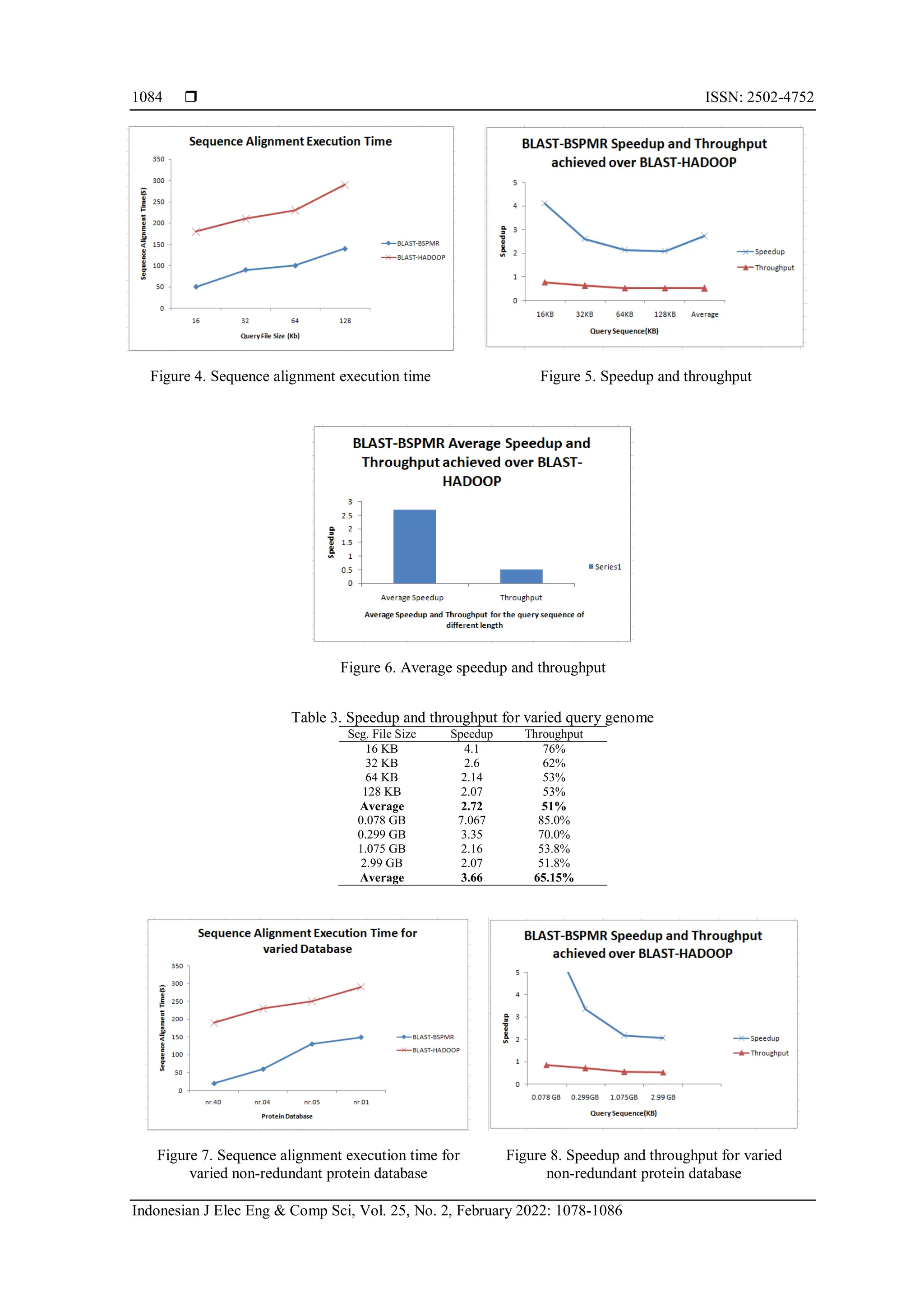
![Indonesian J Elec Eng & Comp Sci ISSN: 2502-4752 An efficient and robust parallel scheduler for bioinformatics … (Leena Ammanna) 1085 Figure 9. Average speedup and throughput non-redundant protein database 4. CONCLUSION Aligning genomic sequences is a straightforward technique for managing and analysing bioinformatics data. The author describes the proper methods for aligning sequences using the BLAST algorithm, which is the most widely used tool for aligning local sequences. Currently, the BLAST algorithm given by NCBI (stand-alone) is unable to handle dynamic biological data in the terabyte range. Cloud platforms are used to address issues such as data storage and data-intensive computation. Biosequencing analysis of genetic data is a critical application. In this study, the author evaluates many existing schedulers. The existing scheduler has issues which are expensive with aligning genomic data sequences. Existing bioinformatics sequence aligners that make use of or embrace Hadoop MapReduce suffer from the concerns discussed in this study. In this paper, the proposed BLAST-BSPMR algorithm is employed to align sequences. For biosequence alignment, the BLASTx algorithm is used, and a parallel MapReduce execution approach based on Azure cloud is used in the BLAST-BSPMR cloud platform. The Map and Reduce framework are executed in parallel to take use of the cloud computing platform's virtual machine-based design. Additionally, the research compares the proposed BLAST-BSPMR technique to previously published system sequence alignments. Experiments are carried out with a variety of non-redundant proteins and nucleotide query sequence files of various sizes. Experiments are presented to demonstrate the efficiency of the BLASTx algorithm. Through an experimental study, a comparison with Hadoop-BLAST for sequence alignment is presented. When comparing the BLAST-BSPMR results to the Hadoop-BLAST results, the BLAST-BSPMR results show a significant improvement. The authors plan to test their theory on a variety of redundant databases in the future such as SwissProt protein, and REFSEQ and also run varied application that are available in NCBI such as BLASTn, and BLASTp on our proposed BSPMR Scheduler to further analyse the robustness and efficacy of our scheduler. REFERENCES [1] D. C. Marinescu, "Parallel and Distributed Computing: Memories of Time Past and a Glimpse at the Future," 2014 IEEE 13th International Symposium on Parallel and Distributed Computing, 2014, pp. 14-15, doi: 10.1109/ISPDC.2014.33. [2] Gartner.Inc, Gartner Forecasts Worldwide Public Cloud End-User Spending to Grow 23% in 2021, 2021. Accessed: Apr. 21, 2021. [Online]. Available: https://www.gartner.com/en/newsroom/press-releases/2021-04-21-gartner-forecasts-worldwide-public- cloud-end-user-spending-to-grow-23-percent-in-2021 [3] W. Hassan, T. S. Chou, T. Omar, and J. Pickard, “Cloud computing survey on services, enhancements and challenges in the era of machine learning and data science,” Int J Inf & Commun Technol., vol. 9, no. 2, pp. 117-139, 2020, doi: 10.11591/ijict.v9i2. [4] W. Hassan, T.-S. Chou, X. Li, P. A.-Kubi, and O. Tamer, “Latest trends, challenges and solutions in security in the era of cloud computing and software defined networks,” Int J Inf & Commun Technol., vol. 8, no. 3, pp. 162-183, 2019, doi: 10.11591/ijict.v8i3.pp162-183. [5] P. Mehrotra, J. Djomegri, S. Heistand, and R. Hood, “Performance evaluation of Amazon EC2 for NASA HPC applications," Proceedings of the 3rd workshop on Scientific Cloud Computing, 2012, pp. 41-50, doi: 10.1145/2287036.2287045. [6] A. Gupta et al., "Evaluating and Improving the Performance and Scheduling of HPC Applications in Cloud," in IEEE Transactions on Cloud Computing, vol. 4, no. 3, pp. 307-321, 1 July-Sept. 2016, doi: 10.1109/TCC.2014.2339858. [7] Z. Cai, X. Li, and J. N. D. Gupta, "Heuristics for Provisioning Services to Workflows in XaaS Clouds," in IEEE Transactions on Services Computing, vol. 9, no. 2, pp. 250-263, 1 March-April 2016, doi: 10.1109/TSC.2014.2361320. [8] M. C. Okur and M. Büyükkeçeci, "Big data challenges in information engineering curriculum," 2014 25th EAEEIE Annual Conference (EAEEIE), 2014, pp. 1-4, doi: 10.1109/EAEEIE.2014.6879372. [9] J. Dean and S. Ghemawat, "MapReduce: simplified data processing on large clusters,” Communications of the ACM, vol. 51, no. 1, pp. 107-113, 2008, doi: 10.1145/1327452.1327492. [10] T. White, Hadoop: The definitive guide, O’Reilly Media, Inc., 2012. [11] A. Hadoop, 2020. Accessed: Sep. 21, 2021. [Online]. Available: http://hadoop.apache.org](https://image.slidesharecdn.com/15440-52181-1-pb-240902031046-71e8d34c/75/An-efficient-and-robust-parallel-scheduler-for-bioinformatics-applications-in-a-public-cloud-A-bigdata-approach-8-2048.jpg)
![ ISSN: 2502-4752 Indonesian J Elec Eng & Comp Sci, Vol. 25, No. 2, February 2022: 1078-1086 1086 [12] J. Zhu, J. Li, E. Hardesty, H. Jiang, and K. Li, "GPU-in-Hadoop: Enabling MapReduce across distributed heterogeneous platforms," 2014 IEEE/ACIS 13th International Conference on Computer and Information Science (ICIS), 2014, pp. 321-326, doi: 10.1109/ICIS.2014.6912154. [13] D. Dahiphale et al., "An Advanced MapReduce: Cloud MapReduce, Enhancements and Applications," in IEEE Transactions on Network and Service Management, vol. 11, no. 1, pp. 101-115, March 2014, doi: 10.1109/TNSM.2014.031714.130407. [14] P. Kondikoppa et al., "MapReduce-Based RESTMD: Enabling Large-Scale Sampling Tasks with Distributed HPC Systems," 2014 6th International Workshop on Science Gateways, 2014, pp. 30-35, doi: 10.1109/IWSG.2014.12. [15] S. Tang, B. Lee, and B. He, "DynamicMR: A Dynamic Slot Allocation Optimization Framework for MapReduce Clusters," in IEEE Transactions on Cloud Computing, vol. 2, no. 3, pp. 333-347, 1 July-Sept. 2014, doi: 10.1109/TCC.2014.2329299. [16] M. Zaharia, A. Konwinski, A. D. Joseph, R. Katz, and I. Stoica "Improving MapReduce performance in heterogeneous environments,” Osdi, vol. 8, no. 4, pp. 29-42, 2008, doi: 10.5555/1855741.1855744. [17] Y. Tao, Q. Zhang, L. Shi, and P. Chen, "Job Scheduling Optimization for Multi-user MapReduce Clusters," 2011 Fourth International Symposium on Parallel Architectures, Algorithms and Programming, 2011, pp. 213-217, doi: 10.1109/PAAP.2011.33. [18] C. Kaushal and D. Koundal, “Recent Trends in Big Data using Hadoop,” International Journal of Informatics and Communication Technology,” International Journal of Informatics and Communication Technology, vol. 8, no. 1, pp. 39-49, 2019, doi: 10.11591/ijict.v8i1.pp39-49. [19] M. Zaharia, D. Borthakur, and J. S. Sarma, "Delay scheduling: a simple technique for achieving locality and fairness in cluster scheduling,” Proceedings of the 5th European conference on Computer systems, 2010, pp. 265-278, doi: 10.1145/1755913.1755940. [20] C. Wang et al., "Mapcheckreduce: an improved mapreduce computing model for imprecise applications,” IEEE International Congress on Big Data, 2014, doi: 10.1109/BigData.Congress.2014.61. [21] G. Malewicz, M. H. Austern, A. J. C. Bik, and J. C. Dehnert, "Pregel: a system for large-scale graph processing,” Proceedings of the 2010 ACM SIGMOD International Conference on Management of data, 2010, pp. 135-146, doi: 10.1145/1807167.1807184. [22] L. G. Valiant, "A bridging model for parallel computation,” Communications of the ACM, vol. 33, no. 8, pp. 103-111, 1990, doi: 10.1145/79173.79181. [23] T. Kajdanowicz, W. Indyk, P. Kazienko, and J. Kukul, "Comparison of the Efficiency of MapReduce and Bulk Synchronous Parallel Approaches to Large Network Processing," 2012 IEEE 12th International Conference on Data Mining Workshops, 2012, pp. 218-225, doi: 10.1109/ICDMW.2012.135. [24] T. Nguyen, W. Shi, and D. Ruden, "CloudAligner: A fast and full-featured MapReduce based tool for sequence mapping,” BMC research notes, vol. 4, no. 1, pp. 1-7, 2011, doi: 10.1186/1756-0500-4-171. [25] M. C. Schatz, “CloudBurst: highly sensitive read mapping with MapReduce,” Bioinformatics, vol. 25, no. 11, 2009, doi: 10.1093/bioinformatics/btp236. BIOGRAPHIES OF AUTHORS Leena Ammanna is currently an Assistant Professor in the department of Information Science and Engineering, SDM College of Engineering and Technology, Dharwad, India. She received B.E. and M.Tech. Degrees from Visvesvaraya Technological University, Karnataka, India. She has 20 years of experience in teaching and research. She can be contacted at leenaignatiusakri@gmail.com. Jagadeeshgowda is a Professor and Head of the Department of Computer Science and Engineering, Shri Krishna Institute of Technology, Bengaluru, India. He has an experience of 24 years in research, academics and industry. He received his Ph.D. from University of JJT,India. Image processing,big data analytics, and network security are his major research interests. He has wide publications in reputed international conferences and journals. He has supervised four Ph.D. and 50 plus Post Graduate students. He has received grants from Government of India. He can be contacted at hodcs.skitblr@gmail.com. Jagadeesh Pujari is a Professor and Head of the Department of Information Science and Engineering, SDM College of Engineering and Technology, Dharwad, India. He has an experience of 32 years in research, academics and industry. He received his Ph.D. from University of Gulbarga, Karnataka, India.His research interests span image processing, pattern recognition, big data analytics, and machine learning. He has wide publications in reputed international conferences and journals. He has supervised five Ph.D. and 80 plus Post Graduate students. He is the investigator for several projects funded by Government of India and Industries. He can be contacted at jaggudp@gmail.com.](https://image.slidesharecdn.com/15440-52181-1-pb-240902031046-71e8d34c/75/An-efficient-and-robust-parallel-scheduler-for-bioinformatics-applications-in-a-public-cloud-A-bigdata-approach-9-2048.jpg)