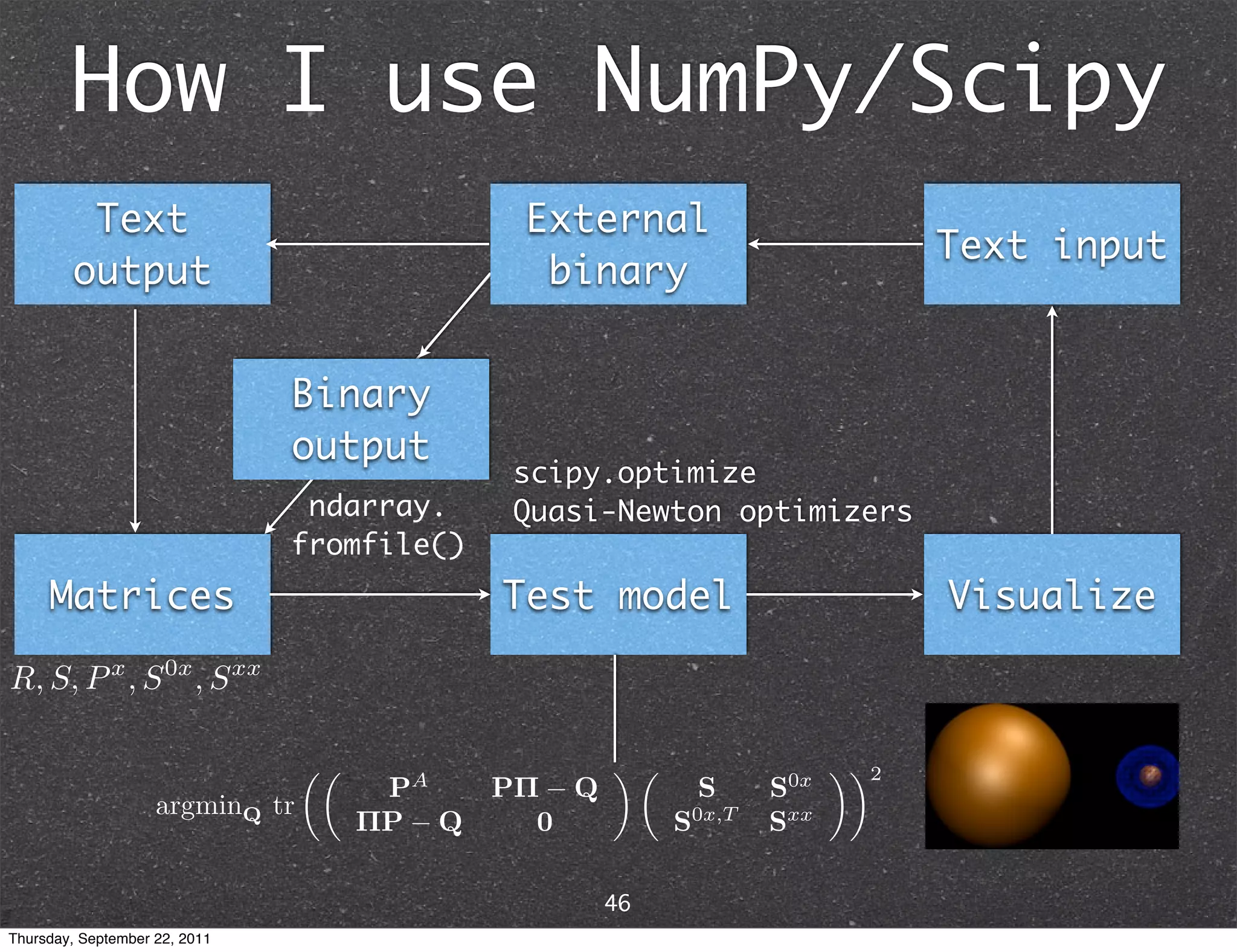
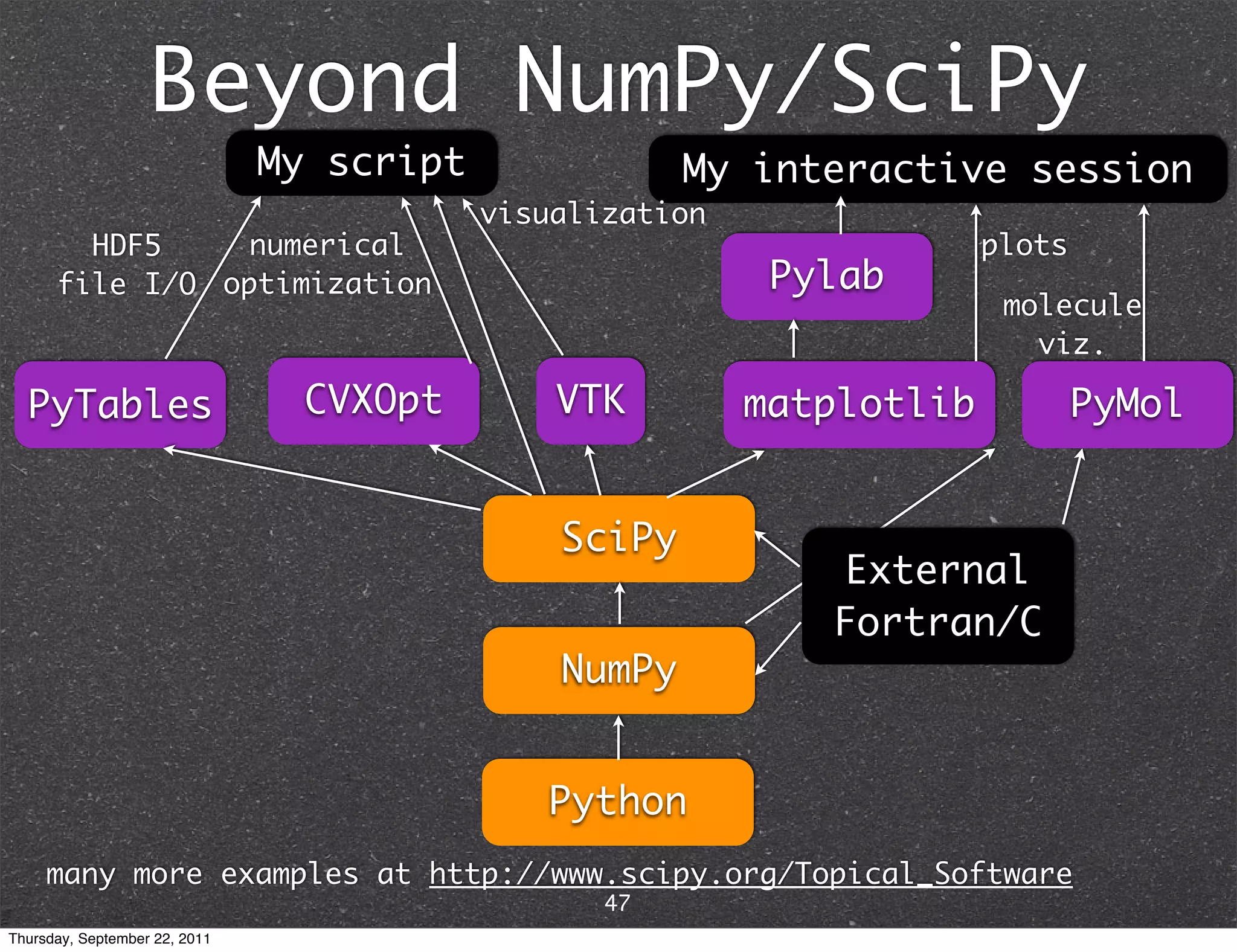
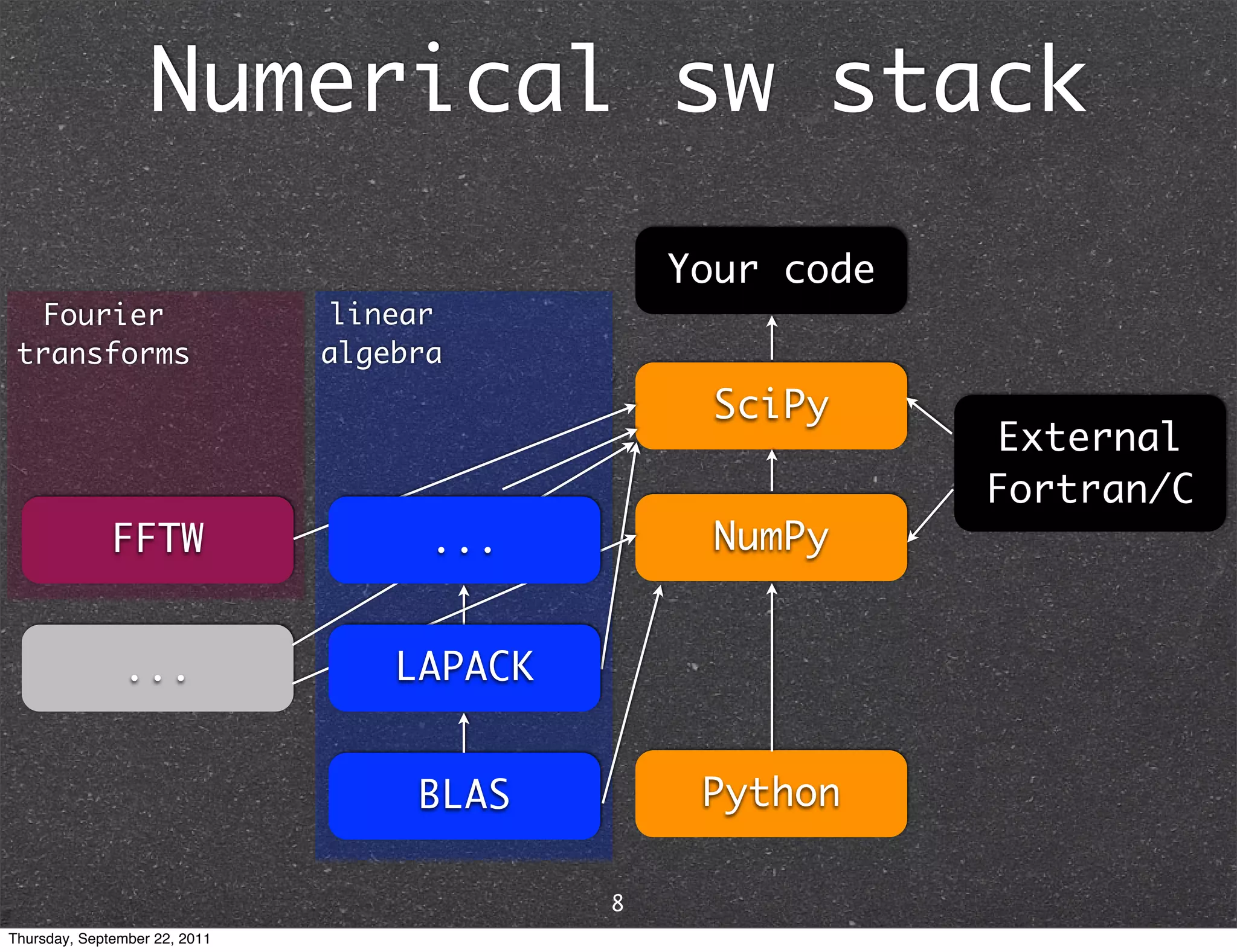
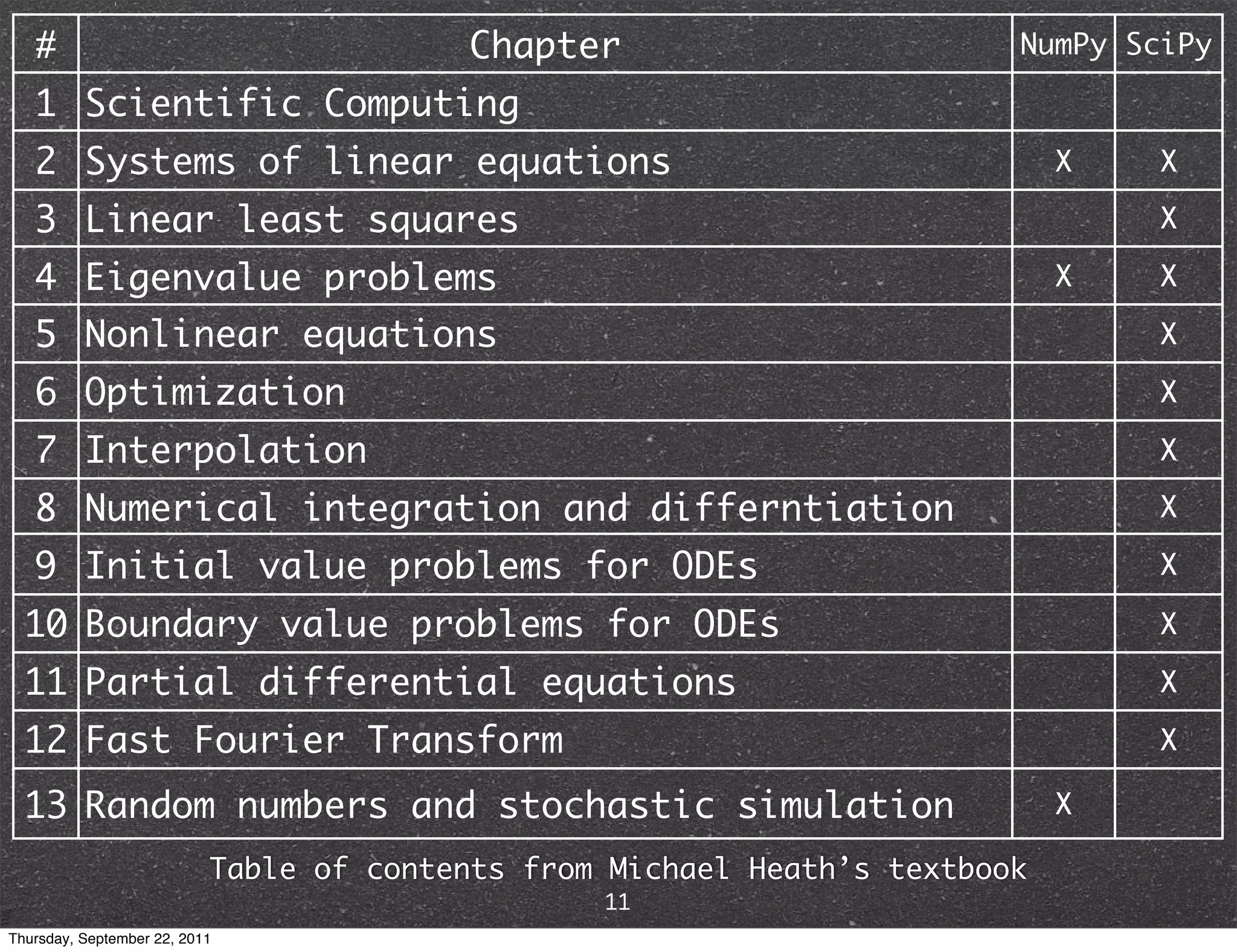
The document discusses NumPy and SciPy, two popular Python packages for scientific computing. NumPy adds support for large, multi-dimensional arrays and matrices to Python. It also introduces data types and affords operations like linear algebra on array objects. SciPy builds on NumPy and contains modules for optimization, integration, interpolation and other tasks. Together, NumPy and SciPy provide a powerful yet easy to use environment for numerical computing in Python.






![Using numpy can make code cleaner import numpy as np a = range(10000000) a = np.arange(10000000) b = range(10000000) b = np.arange(10000000) c = [] c = a + b for i in range(len(a)): c.append(a[i] + b[i]) What’s different?? 6 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-7-2048.jpg)
![What’s different?creates list of creates ndarray of dynamically typed int statically typed int import numpy as np a = range(10000000) a = np.arange(10000000) b = range(10000000) b = np.arange(10000000) c = [] #a+b is concatenation c = a + b #vectorized addition for i in range(len(a)): c.append(a[i] + b[i]) Using numpy can save lots of time 7.050s 0.333s (21x) a convenient interface to compiled C/Fortran libraries: BLAS, LAPACK, FFTW, UMFPACK,... 7 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-8-2048.jpg)


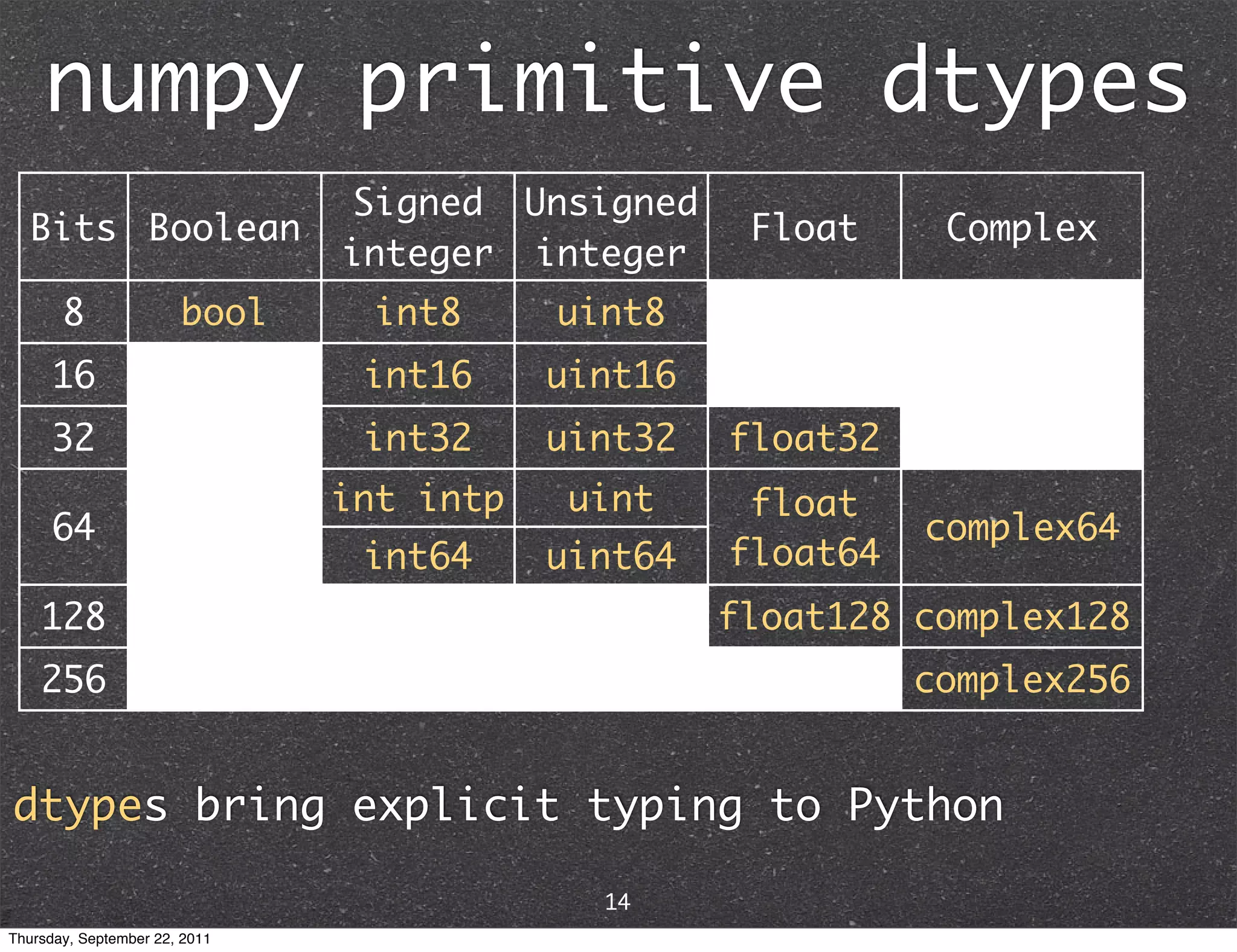
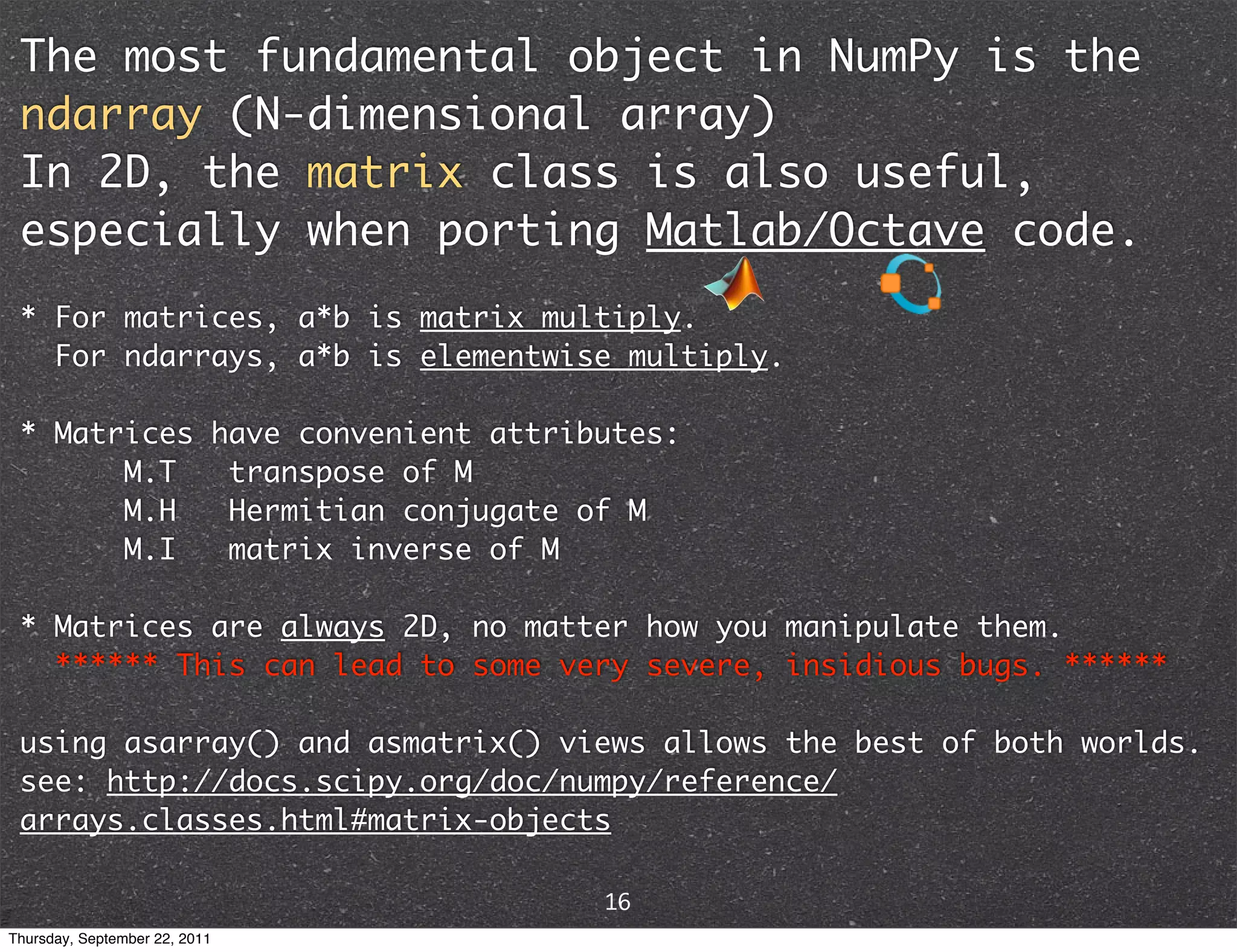
![The most fundamental object in NumPy is the ndarray (N-dimensional array) v[:] vector M[:,:] matrix x[:,:,...,:] higher order tensor unlike built-in Python data types, ndarrays are designed for homogeneous, explicitly typed data 13 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-15-2048.jpg)
![Structured array: ndarray of data structure >>> mol = np.zeros(3, dtype=('uint8, 3float64')) >>> mol[0] = 8, (-0.464, 0.177, 0.0) >>> mol[1] = 1, (-0.464, 1.137, 0.0) >>> mol[2] = 1, (0.441, -0.143, 0.0) >>> mol array([(8, [-0.46400000000000002, 0.17699999999999999, 0.0]), (1, [-0.46400000000000002, 1.137, 0.0]), (1, [0.441, -0.14299999999999999, 0.0])], dtype=[('f0', '|u1'), ('f1', '<f8', (3,))]) Recarray: ndarray of data structure with named fields (record) >>> mol = np.array(mol, dtype={'atomicnum':('uint8',0), 'coords':('3float64',1)}) >>> mol['atomicnum'] array([8, 1, 1], dtype=uint8) 15 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-17-2048.jpg)
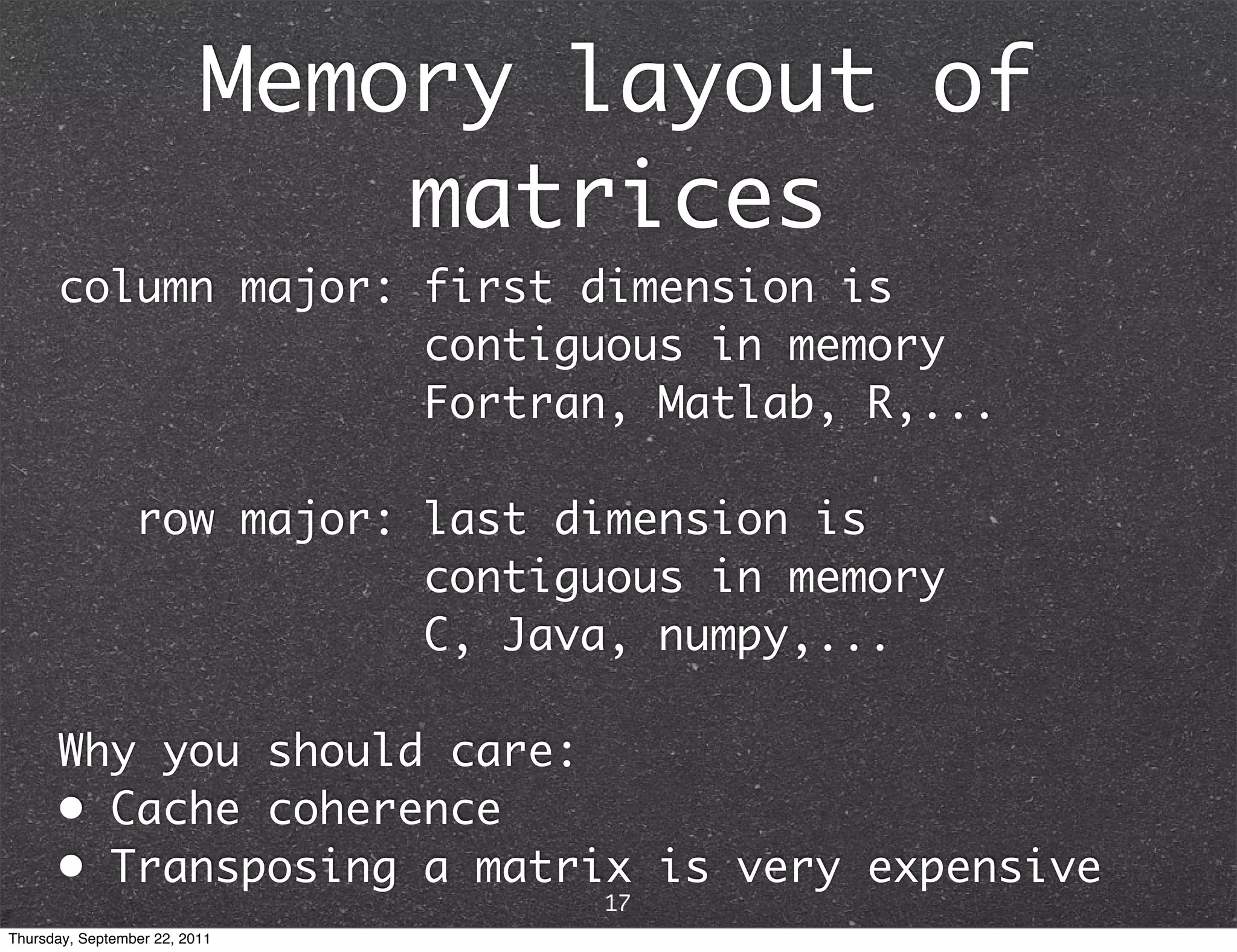
![Creating ndarrays • from Python iterable: lists, tuples,... e.g. array([1, 2, 3]) == asarray((1, 2, 3)) • from intrinsic functions empty() allocates memory only zeros() initializes to 0 ones() initializes to 1 arange() creates a uniform range rand() initializes to uniform random randn() initializes to standard normal random ... • from binary representation in string/buffer • from file on disk 18 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-20-2048.jpg)
![Generating ndarrays fromfunction() creates an ndarray whose entries are functions of its indices e.g. the Hilbert matrix 1..n >>> np.fromfunction(lambda i,j: 1./(i+j+1), (4,4)) array([[ 1. , 0.5 , 0.33333333, 0.25 ], [ 0.5 , 0.33333333, 0.25 , 0.2 ], [ 0.33333333, 0.25 , 0.2 , 0.16666667], [ 0.25 , 0.2 , 0.16666667, 0.14285714]]) 19 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-21-2048.jpg)
![Generating ndarrays arange(): like range() but accepts floats >>> import numpy as np >>> np.arange(2, 2.5, 0.1) array([ 2. , 2.1, 2.2, 2.3, 2.4]) linspace(): creates array with specified number of elements, spaced equally between the specified beginning and ending. >>> np.linspace(2.0, 2.4, 5) array([ 2. , 2.1, 2.2, 2.3, 2.4]) 20 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-22-2048.jpg)
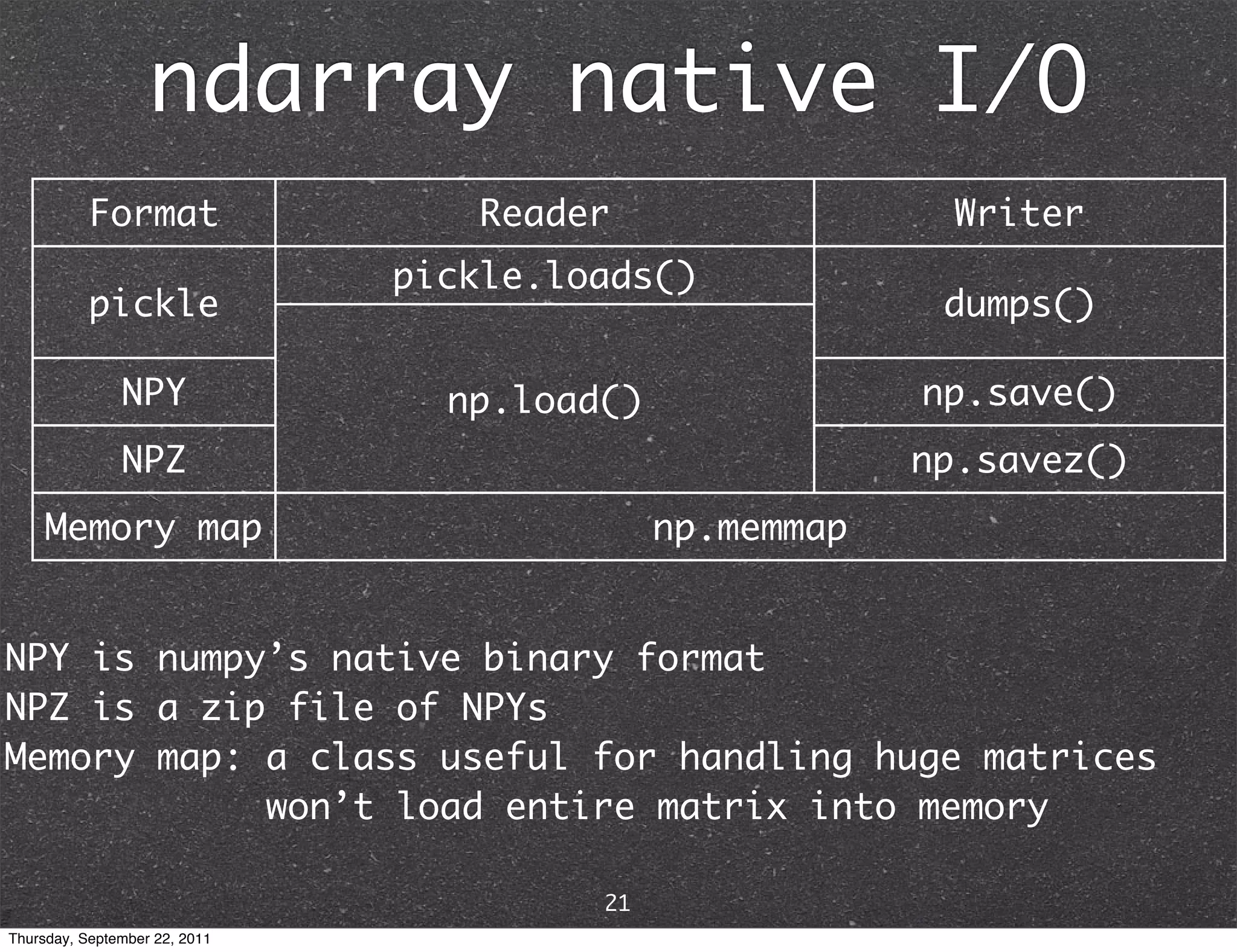

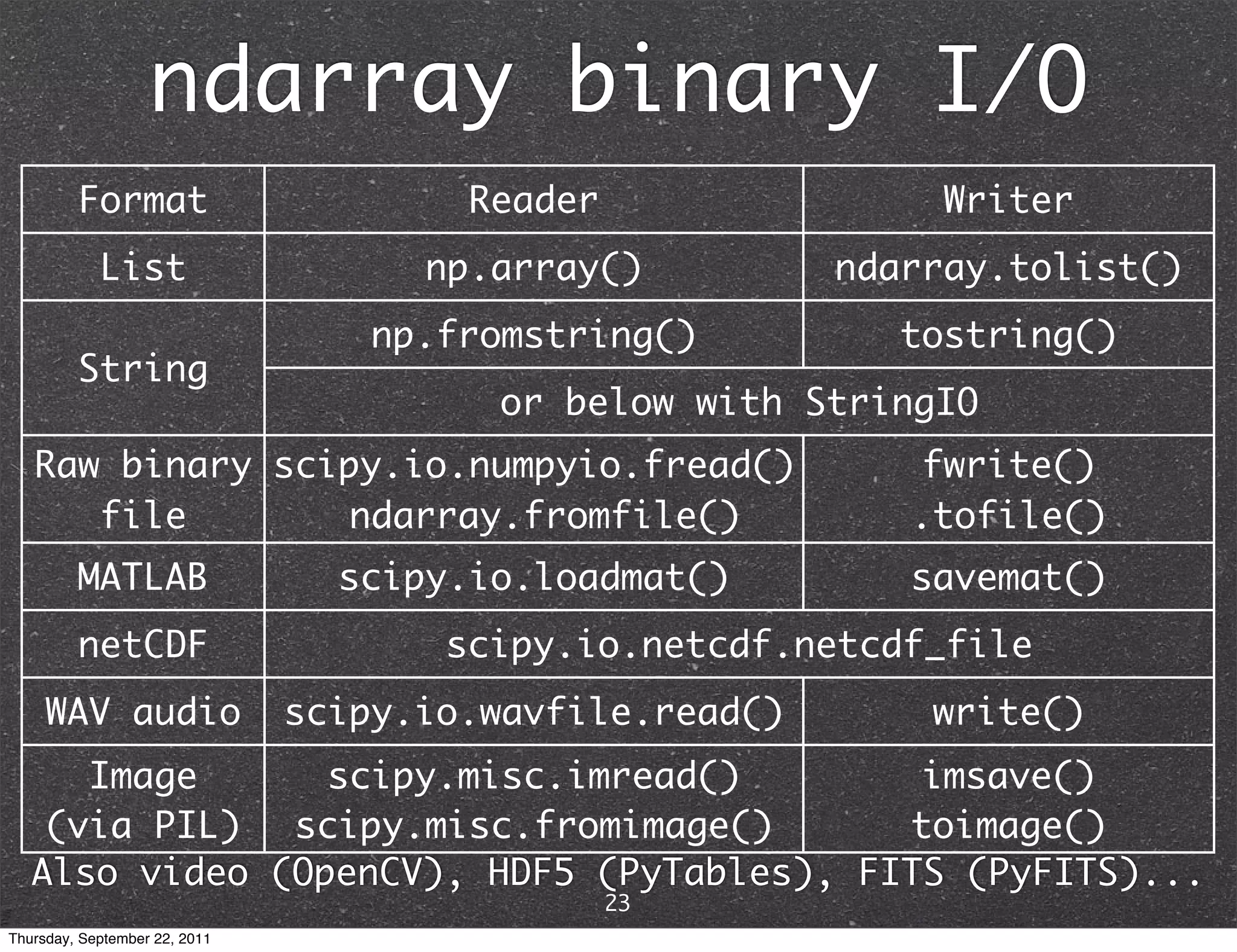
![Indexing >>> x = np.arange(12).reshape(3,4); x array([[ 0, 1, 2, 3], [ 4, 5, 6, 7], [ 8, 9, 10, 11]]) >>> x[1,2] 6 >>> x[2,-1] row, then column 11 >>> x[0][2] 2 >>> x[(2,2)] #tuple 10 >>> x[:1] #slices return views, not copies array([[0, 1, 2, 3]]) >>> x[::2,1:4:2] array([[ 1, 3], [ 9, 11]]) 24 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-26-2048.jpg)
![Fancy indexing >>> x = np.arange(12).reshape(3,4); x array([[ 0, 1, 2, 3], [ 4, 5, 6, 7], [ 8, 9, 10, 11]]) >>> x[(2,2)] array index 10 >>> x[np.array([2,2])] #same as x[[2,2]] array([[ 8, 9, 10, 11], [ 8, 9, 10, 11]]) >>> x[np.array([1,0]), np.array([2,1])] array([6, 1]) >>> x[x>8] Boolean mask array([ 9, 10, 11]) >>> x>8 array([[False, False, False, False], [False, False, False, False], [False, True, True, True]], dtype=bool) 25 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-27-2048.jpg)
![Fancy indexing II >>> y = np.arange(1*2*3*4).reshape(1,2,3,4); y array([[[[ 0, 1, 2, 3], [ 4, 5, 6, 7], [ 8, 9, 10, 11]], [[12, 13, 14, 15], [16, 17, 18, 19], [20, 21, 22, 23]]]]) >>> y[0, Ellipsis, 0] # == y[0, ..., 0] == [0,:,:,0] array([[ 0, 4, 8], [12, 16, 20]]) >>> y[0, 0, 0, slice(2,4)] # == y[(0, 0, 0, 2:4)] array([2, 3]) 26 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-28-2048.jpg)
![Broadcasting What happens when you multiply ndarrays of different dimensions? Case I: trailing dimensions match >>> x #.shape = (3,4) array([[ 0, 1, 2, 3], >>> y * x [ 4, 5, 6, 7], array([[[[ 0, 1, 4, 9], [ 8, 9, 10, 11]]) [ 16, 25, 36, 49], >>> y #.shape = (1,2,3,4) [ 64, 81, 100, 121]], array([[[[ 0, 1, 2, 3], [ 4, 5, 6, 7], [[ 0, 13, 28, 45], [ 8, 9, 10, 11]], [ 64, 85, 108, 133], [160, 189, 220, 253]]]]) [[12, 13, 14, 15], [16, 17, 18, 19], [20, 21, 22, 23]]]]) 27 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-29-2048.jpg)
![Broadcasting What happens when you multiply ndarrays of different dimensions? Case II: trailing dimension is 1 >>> b.shape = 4,1 >>> a + b >>> a = np.arange(4); a array([[3, 4, 5, 6], array([0, 1, 2, 3]) [2, 3, 4, 5], >>> b = np.arange(4)[::-1]; b [1, 2, 3, 4], array([3, 2, 1, 0]) [0, 1, 2, 3]]) >>> a + b array([3, 3, 3, 3]) >>> b.shape = 1,4 >>> a + b array([[3, 3, 3, 3]]) 28 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-30-2048.jpg)

![Matrix functions You can apply a function elementwise to a matrix... >>> from numpy import array, exp >>> X = array([[1, 1], [1, 0]]) >>> exp(X) array([[ 2.71828183, 2.71828183], [ 2.71828183, 1.]]) ...or a matrix version of that function >>> from scipy.linalg import expm >>> expm(X) array([[ 2.71828183, 7.3890561 ], [ 1. , 2.71828183]]) other functions in scipy.linalg.matfuncs 30 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-32-2048.jpg)

![Least-squares curve fitting from scipy import * fit data to model from scipy.optimize import leastsq from matplotlib.pyplot import plot #Make up data x(t) with Gaussian noise num_points = 150 t = linspace(5, 8, num_points) x = 11.86*cos(2*pi/0.81*t-1.32) + 0.64*t +4*((0.5-rand(num_points))* exp(2*rand(num_points)**2)) # Target function model = lambda p, x: p[0]*cos(2*pi/p[1]*x+p[2]) + p[3]*x # Distance to the target function error = lambda p, x, y: model(p, x) - y # Initial guess for the parameters p0 = [-15., 0.8, 0., -1.] p1, _ = leastsq(error, p0, args=(t, x)) t2 = linspace(t.min(), t.max(), 100) plot(t, x, "ro", t2, model(p1, t2), "b-") raw_input() 32 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-34-2048.jpg)



![From math to code import numpy #Make up some data N = 30000 idx = 24700 size = 300 data = numpy.random.rand(N) frag_pad = numpy.zeros(N) frag = data[idx:idx+size] frag_pad[:size] = frag #Compute phase correlation data_ft = numpy.fft.rfft(data) frag_ft = numpy.fft.rfft(frag_pad) phase = data_ft * numpy.conj(frag_ft) phase /= abs(phase) cross_correlation = numpy.fft.irfft(phase) offset = numpy.argmax(cross_correlation) print 'Input offset: %d, computed: %d' % (idx, offset) from matplotlib.pyplot import plot plot(cross_correlation) raw_input() #Pause 35 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-38-2048.jpg)
![From math to code import numpy #Make up some data N = 30000 idx = 24700 size = 300 data = numpy.random.rand(N) frag_pad = numpy.zeros(N) frag = data[idx:idx+size] frag_pad[:size] = frag #Compute phase correlation data_ft = numpy.fft.rfft(data) frag_ft = numpy.fft.rfft(frag_pad) phase = data_ft * numpy.conj(frag_ft) phase /= abs(phase) cross_correlation = numpy.fft.irfft(phase) offset = numpy.argmax(cross_correlation) print 'Input offset: %d, computed: %d' % (idx, offset) from matplotlib.pyplot import plot plot(cross_correlation) raw_input() #Pause 35 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-39-2048.jpg)


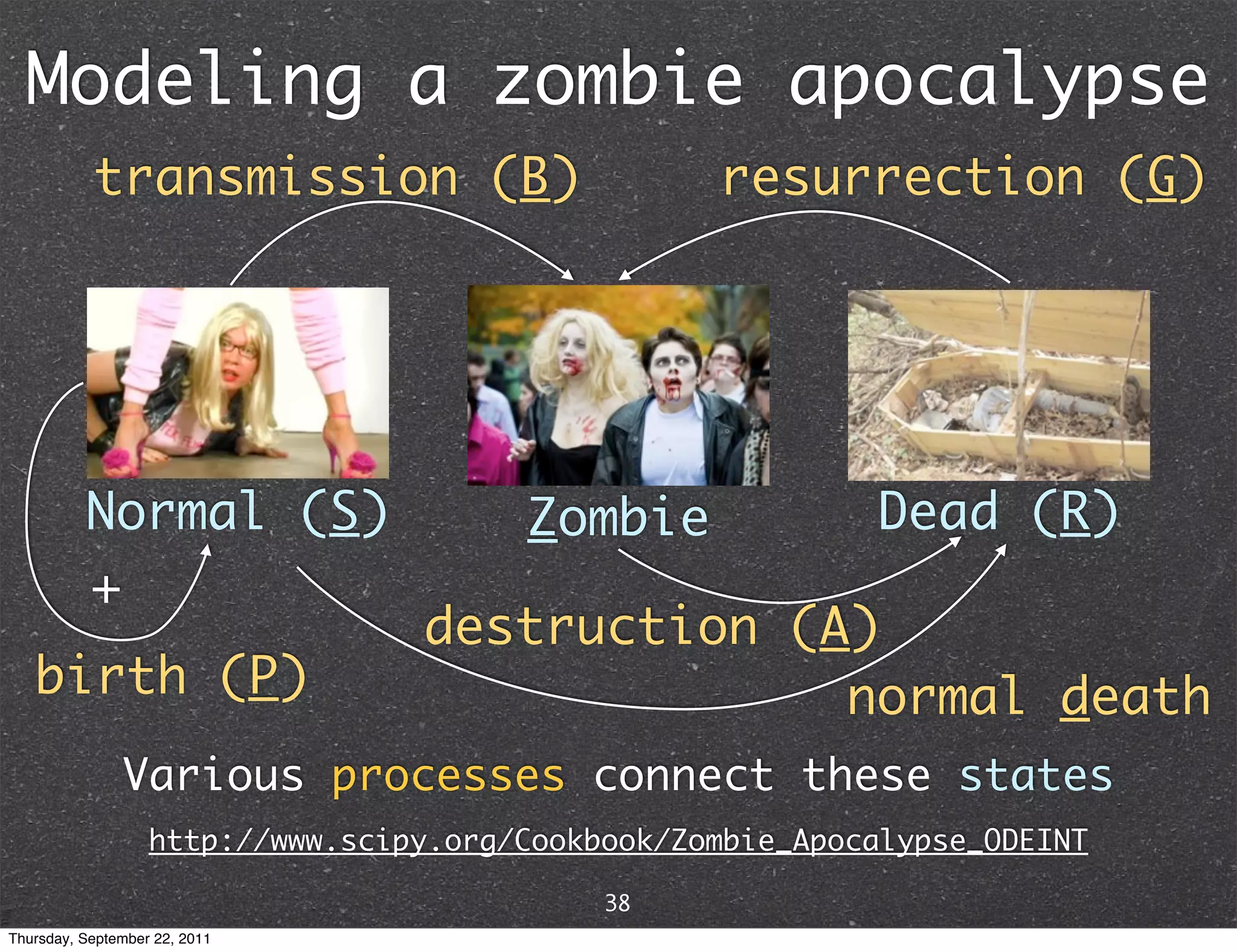
![From math to code B G from numpy import linspace from scipy.integrate import odeint P = 0 # birth rate d = 0.0001 # natural death rate S Z R B = 0.0095 # transmission rate + A G = 0.0001 # resurrection rate A = 0.0001 # destruction rate r d def f(y, t): Si, Zi, Ri = y return [P - B*Si*Zi - d*Si, B*Si*Zi + G*Ri - A*Si*Zi, d*Si + A*Si*Zi - G*Ri] y0 = [500, 0, 0] # initial conditions t = linspace(0, 5., 1000) # time grid soln = odeint(f, y0, t) # solve ODE S, Z, R = soln[:, :].T http://www.scipy.org/Cookbook/Zombie_Apocalypse_ODEINT 39 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-43-2048.jpg)


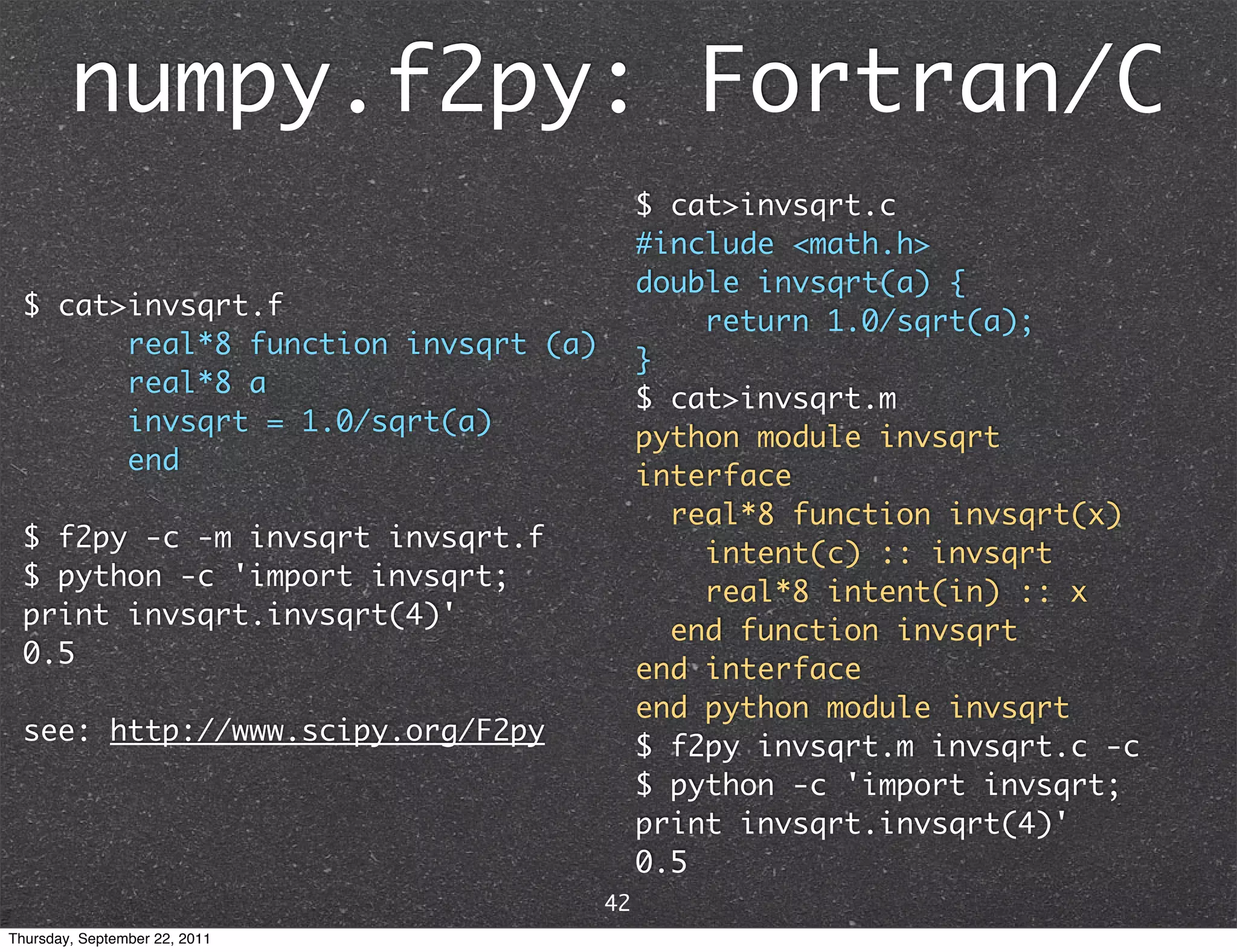
![scipy.weave.inline python on-the-fly scipy distutils compiled weave core C/C++ program inline Extension >>> from scipy.weave import inline >>> x = 4.0 >>> inline('return_val = 1./sqrt(x));', ['x']) 0.5 see: https://github.com/scipy/scipy/blob/ master/scipy/weave/doc/tutorial.txt 43 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-47-2048.jpg)
![scipy.weave.blitz Uses the Blitz++ numerical library for C++ Converts between ndarrays and Blitz arrays >>> # Computes five-point average using numpy and weave.blitz >>> import numpy import empty >>> from scipy.weave import blitz >>> a = numpy.zeros((4096,4096)); c = numpy.zeros((4096, 4096)) >>> b = numpy.random.randn(4096,4096) >>> c[1:-1,1:-1] = (b[1:-1,1:-1] + b[2:,1:-1] + b[:-2,1:-1] + b [1:-1,2:] + b[1:-1,:-2]) / 5.0 >>> blitz("a[1:-1,1:-1] = (b[1:-1,1:-1] + b[2:,1:-1] + b [:-2,1:-1] + b[1:-1,2:] + b[1:-1,:-2]) / 5.") >>> (a == c).all() True see: https://github.com/scipy/scipy/blob/master/scipy/weave/doc/ tutorial.txt 44 Thursday, September 22, 2011](https://image.slidesharecdn.com/numpyscipy-110922225541-phpapp01/75/Python-as-number-crunching-code-glue-48-2048.jpg)