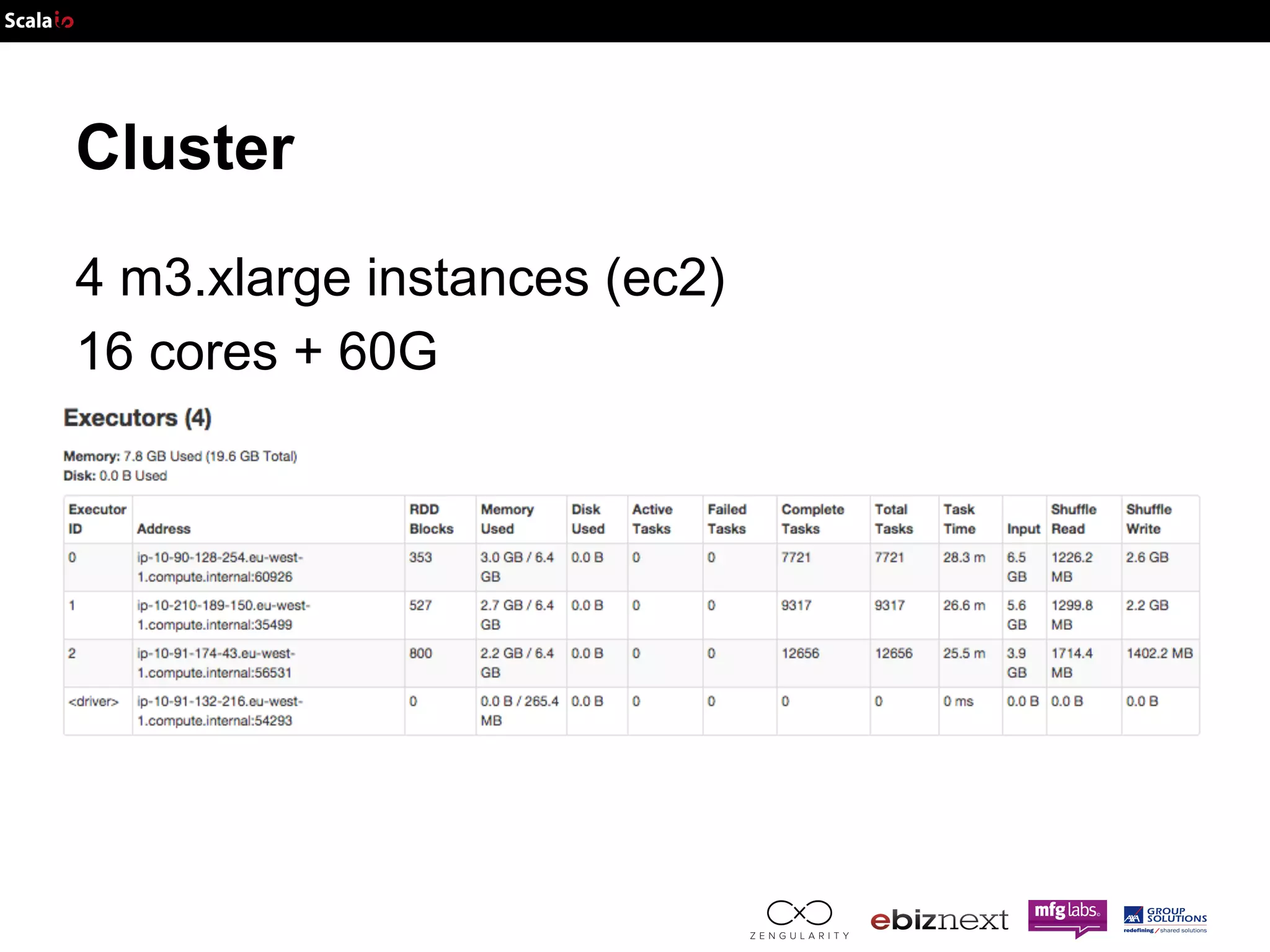
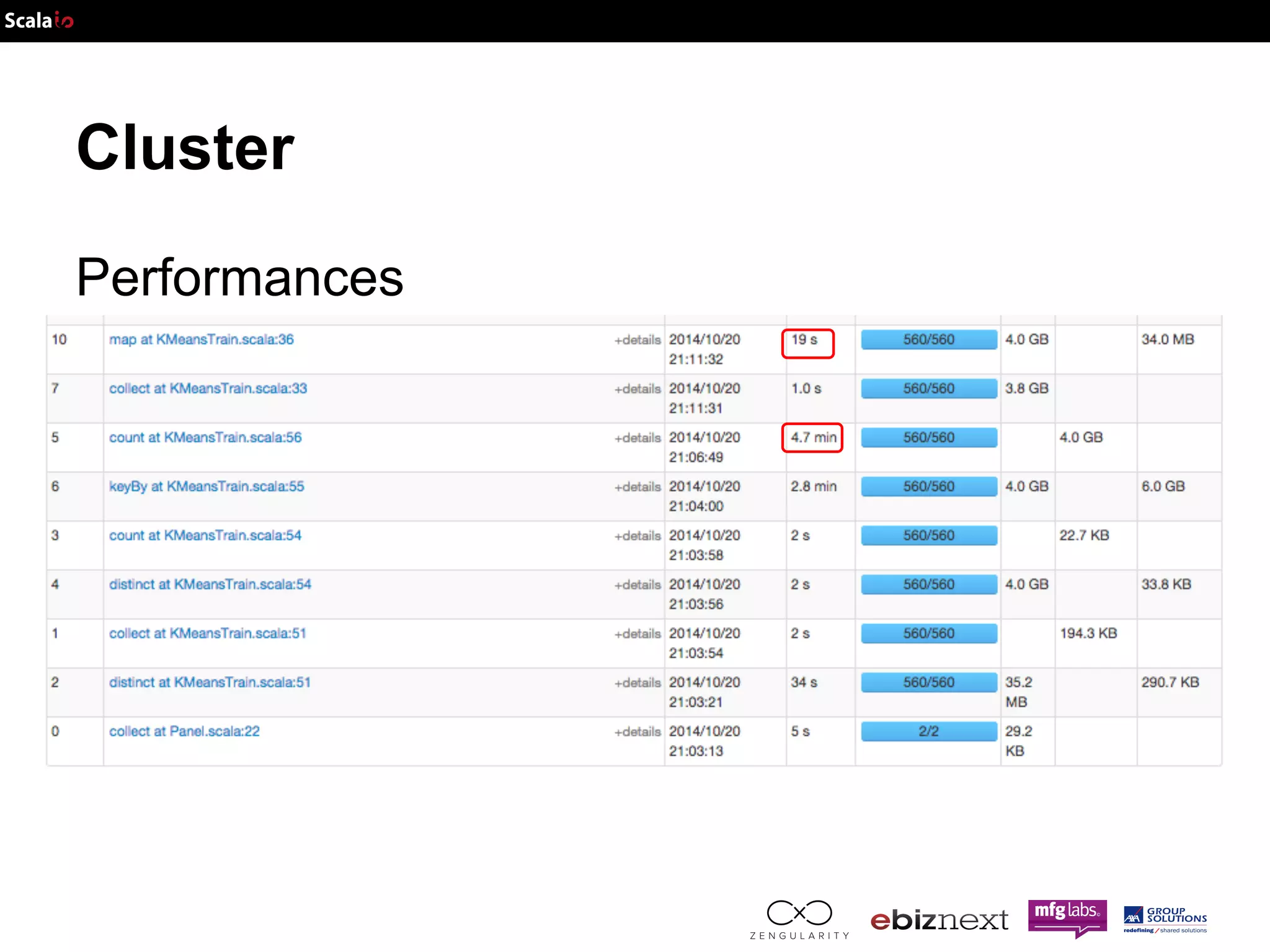
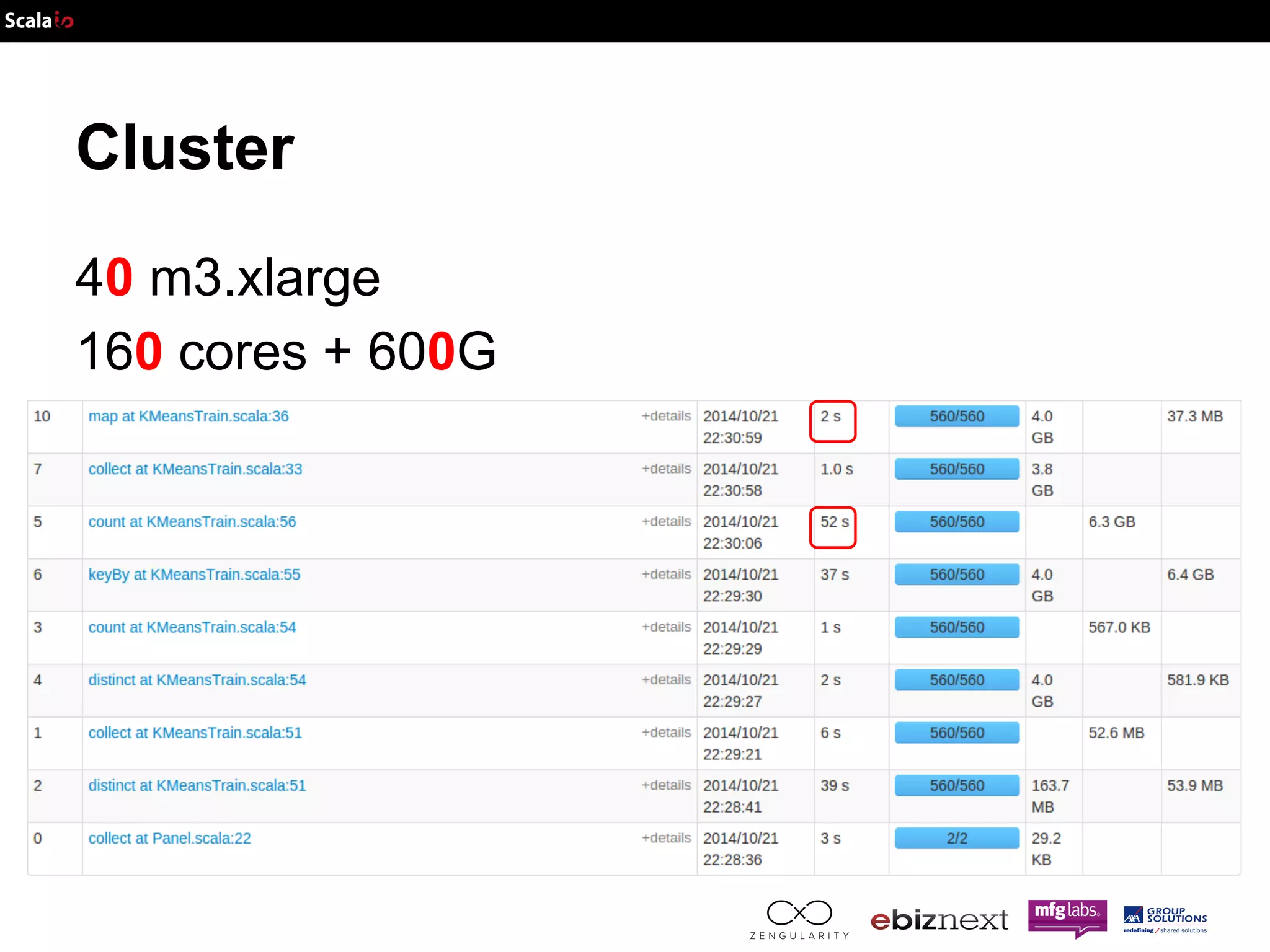
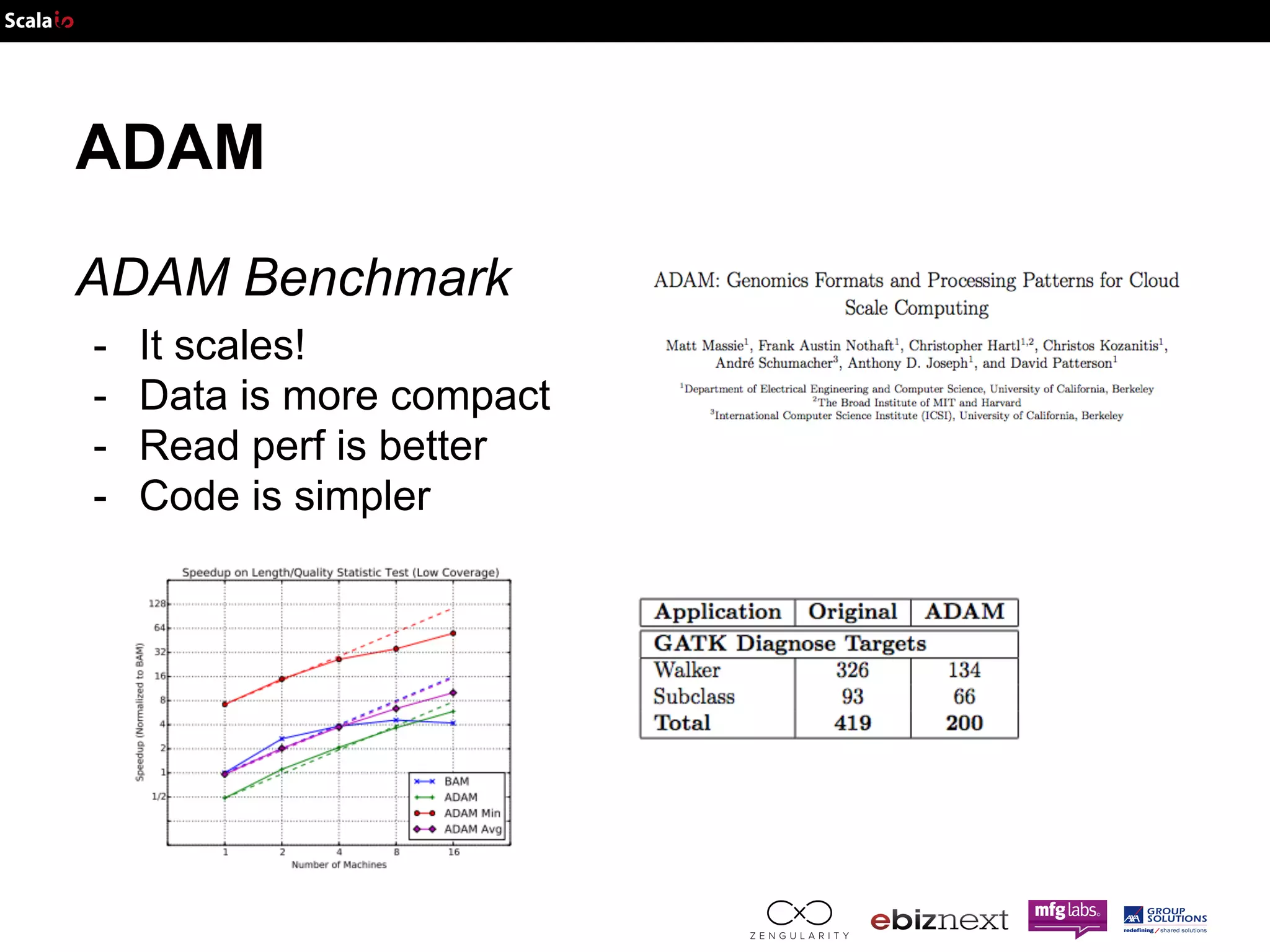
This document discusses using Apache Spark and ADAM to perform scalable genomic analysis. It provides an overview of genomics and challenges with existing approaches. ADAM uses Apache Spark and Parquet to efficiently store and query large genomic datasets. The document demonstrates clustering genomic data from the 1000 Genomes Project to predict populations, showing ADAM and Spark can handle large genomic workloads. It concludes these tools provide scalable genomic data processing but future work is needed to implement more advanced algorithms.




![Parquet 101 Efficient (schema based) serialization: AVRO JSON Schema IDL { "namespace": "example.avro", "type": "record", "name": "User", "fields": [ {"name": "name", "type": "string"}, {"name": "favorite_number", "type": ["int", "null"]}, {"name": "favorite_color", "type": ["string", "null"]} ] } record User { string name; union { null, int } favorite_number = null; union { null, string } favorite_color = null; }](https://image.slidesharecdn.com/adam-141025105017-conversion-gate02/75/Lightning-fast-genomics-with-Spark-Adam-and-Scala-16-2048.jpg)
![Parquet 101 Efficient (schema based) serialization: AVRO JSON Schema Part of the: { "namespace": "example.avro", "type": "record", "name": "User", "fields": [ {"name": "name", "type": "string"}, {"name": "favorite_number", "type": ["int", "null"]}, {"name": "favorite_color", "type": ["string", "null"]} ] } ● protocol ● serialization →less metadata Define: IDL → JSON Send: Binary → JSON](https://image.slidesharecdn.com/adam-141025105017-conversion-gate02/75/Lightning-fast-genomics-with-Spark-Adam-and-Scala-17-2048.jpg)
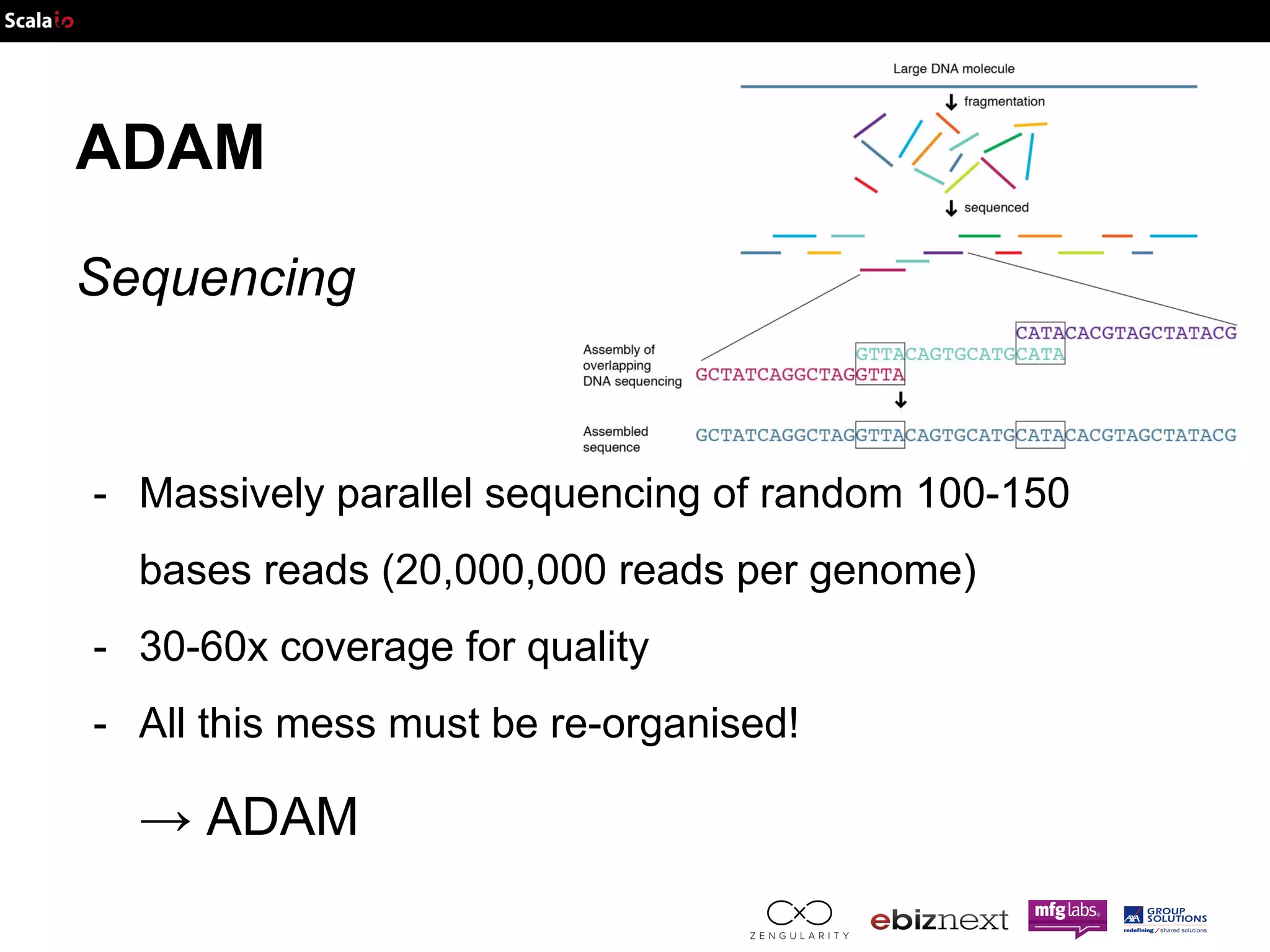
![ADAM Overview (Sequencing) - DNA is a molecule …or a Seq[Char] (A, T, G, C) alphabet](https://image.slidesharecdn.com/adam-141025105017-conversion-gate02/75/Lightning-fast-genomics-with-Spark-Adam-and-Scala-19-2048.jpg)

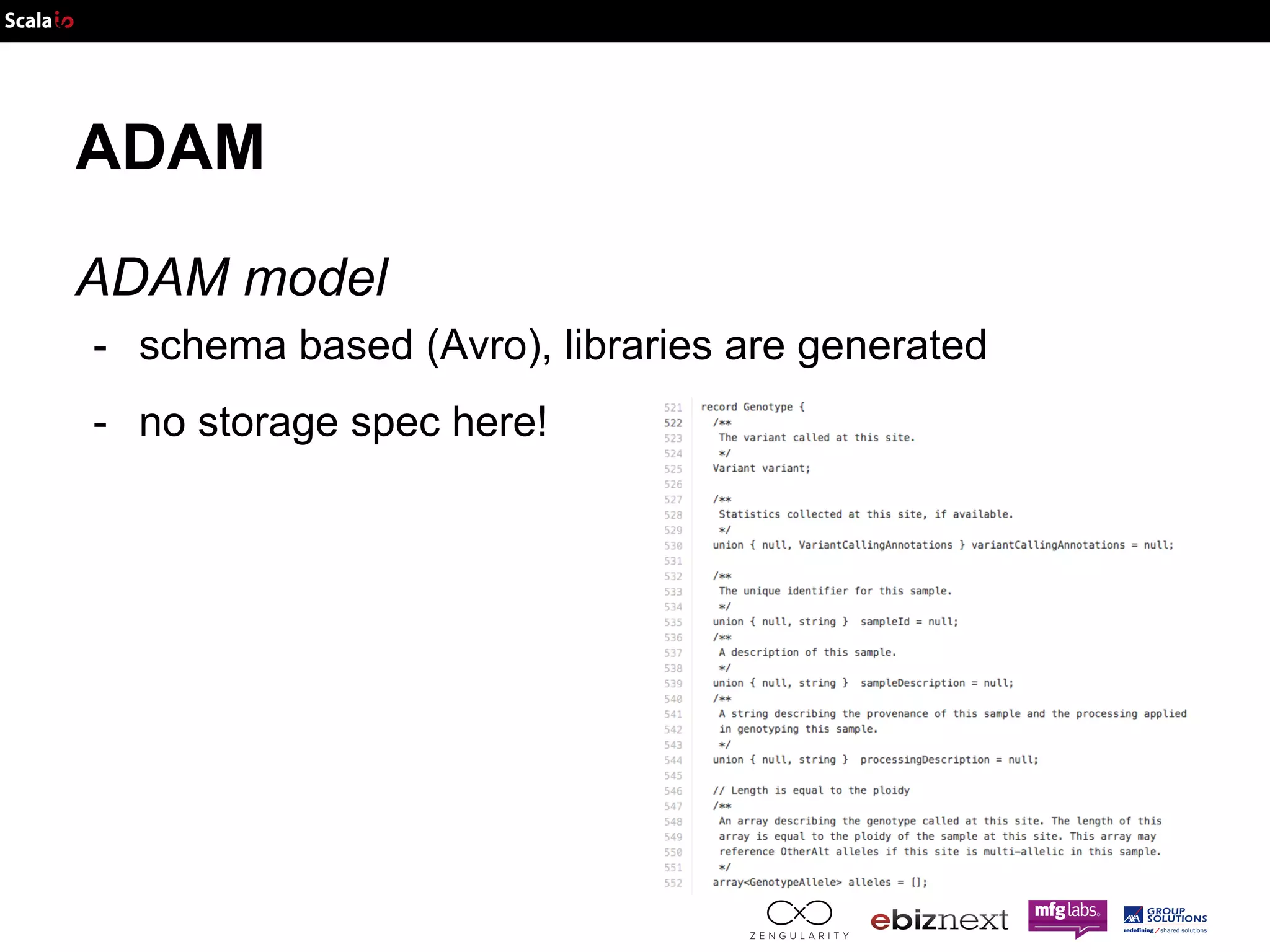



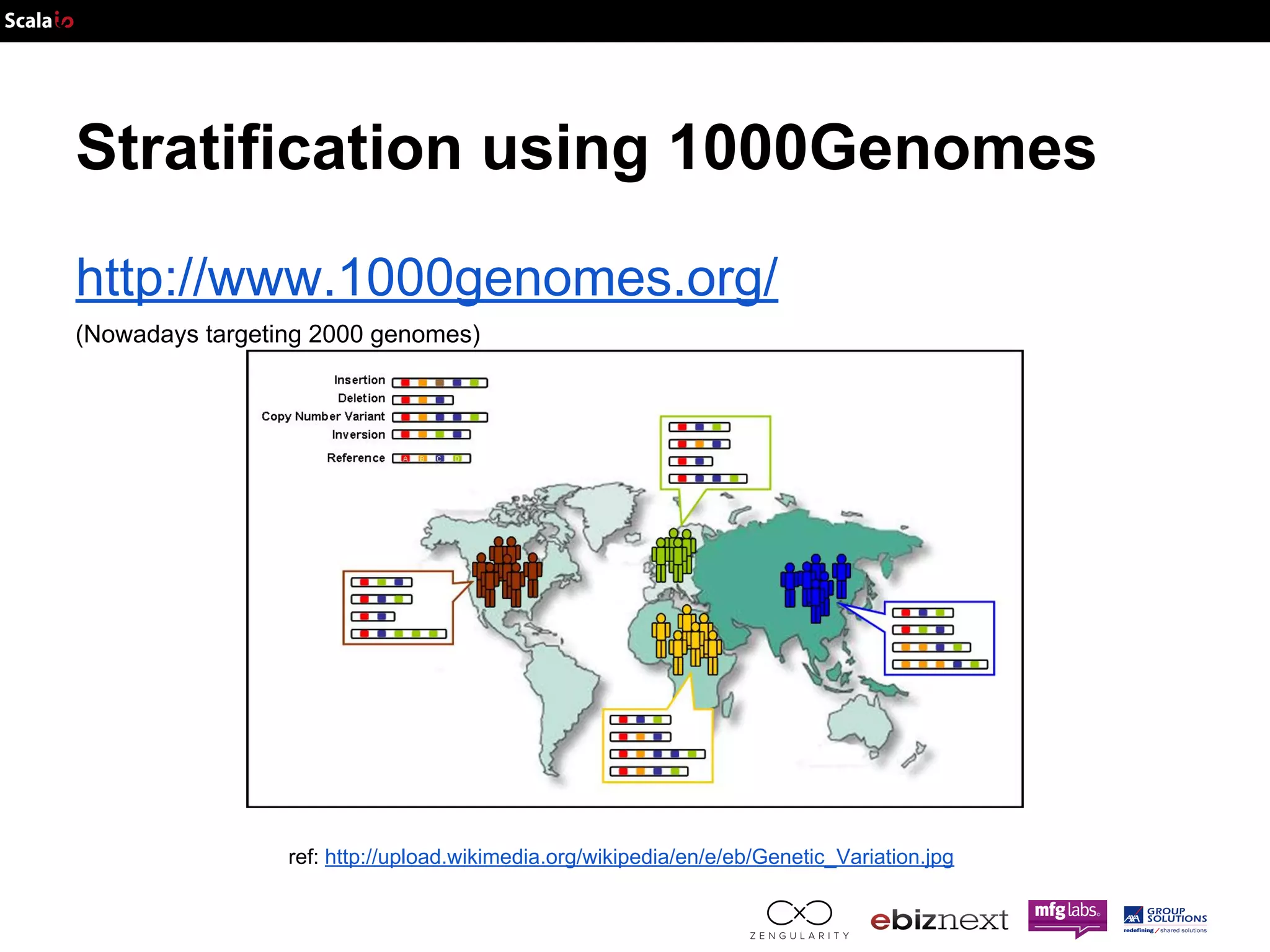


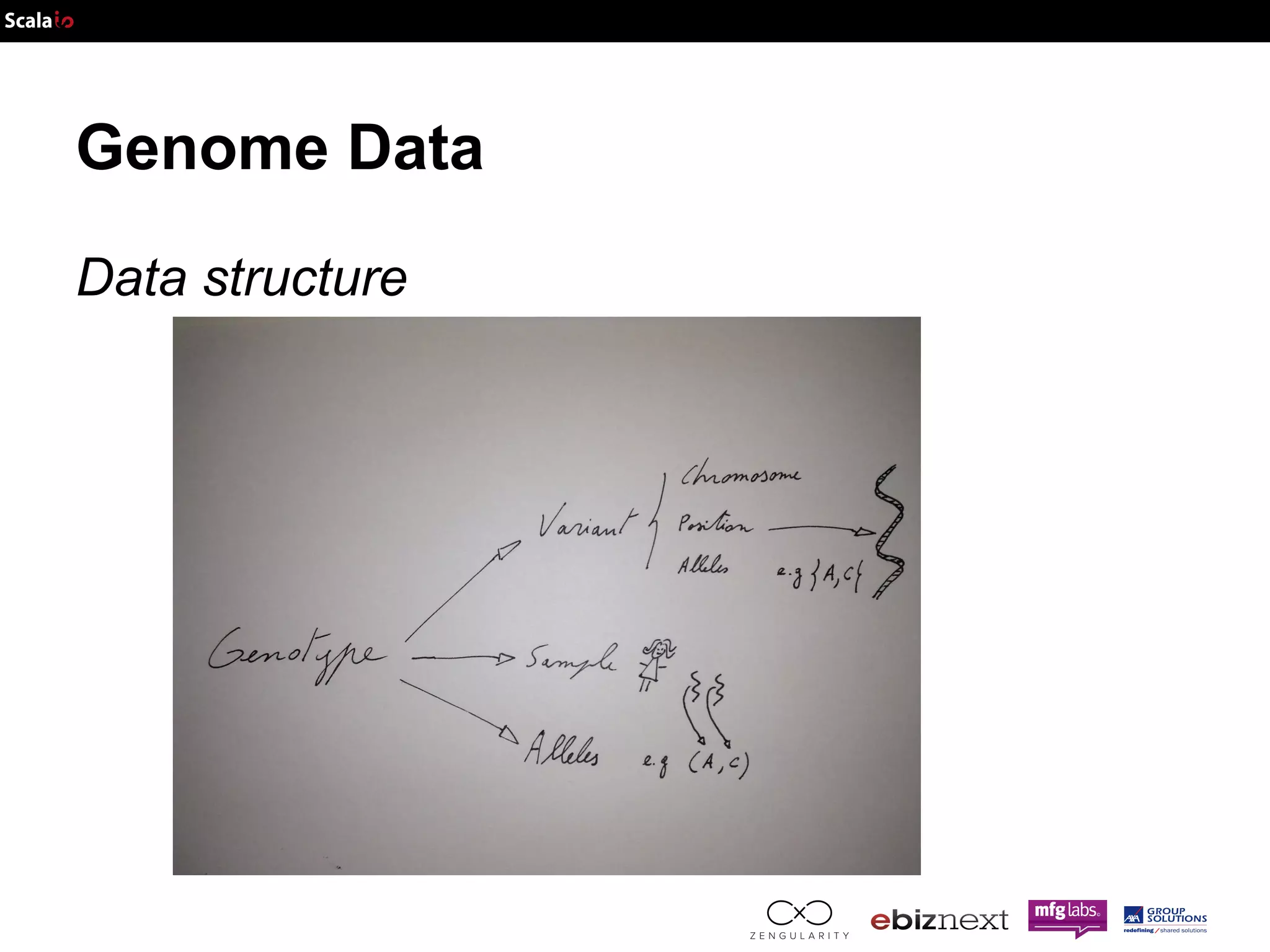
![Genome Data Data structure Panel: Map[SampleID, Population]](https://image.slidesharecdn.com/adam-141025105017-conversion-gate02/75/Lightning-fast-genomics-with-Spark-Adam-and-Scala-37-2048.jpg)

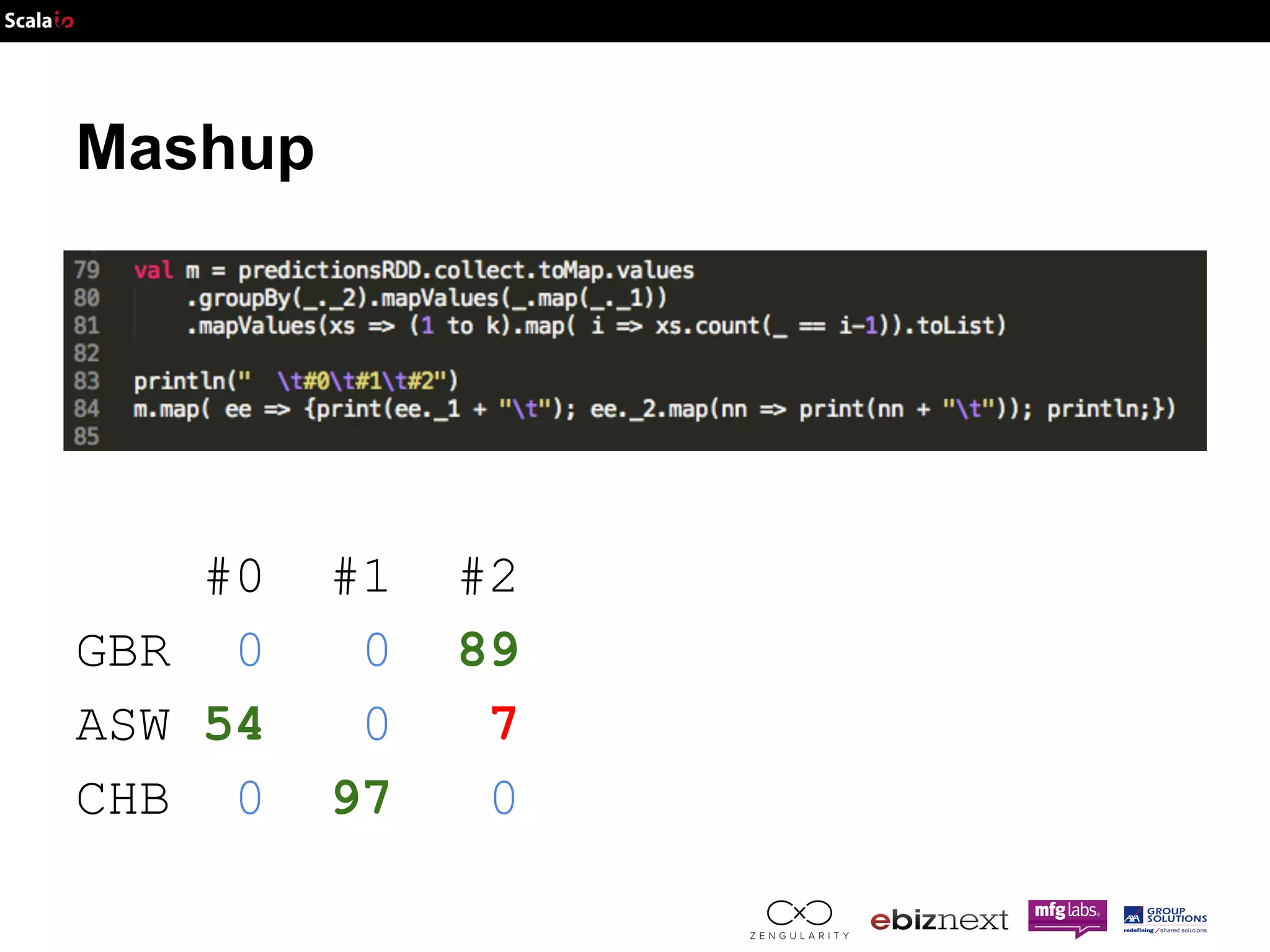
![Mashup prediction Sample [NA20332] is in cluster #0 for population Some(ASW) Sample [NA20334] is in cluster #2 for population Some(ASW) Sample [HG00120] is in cluster #2 for population Some(GBR) Sample [NA18560] is in cluster #1 for population Some(CHB)](https://image.slidesharecdn.com/adam-141025105017-conversion-gate02/75/Lightning-fast-genomics-with-Spark-Adam-and-Scala-43-2048.jpg)