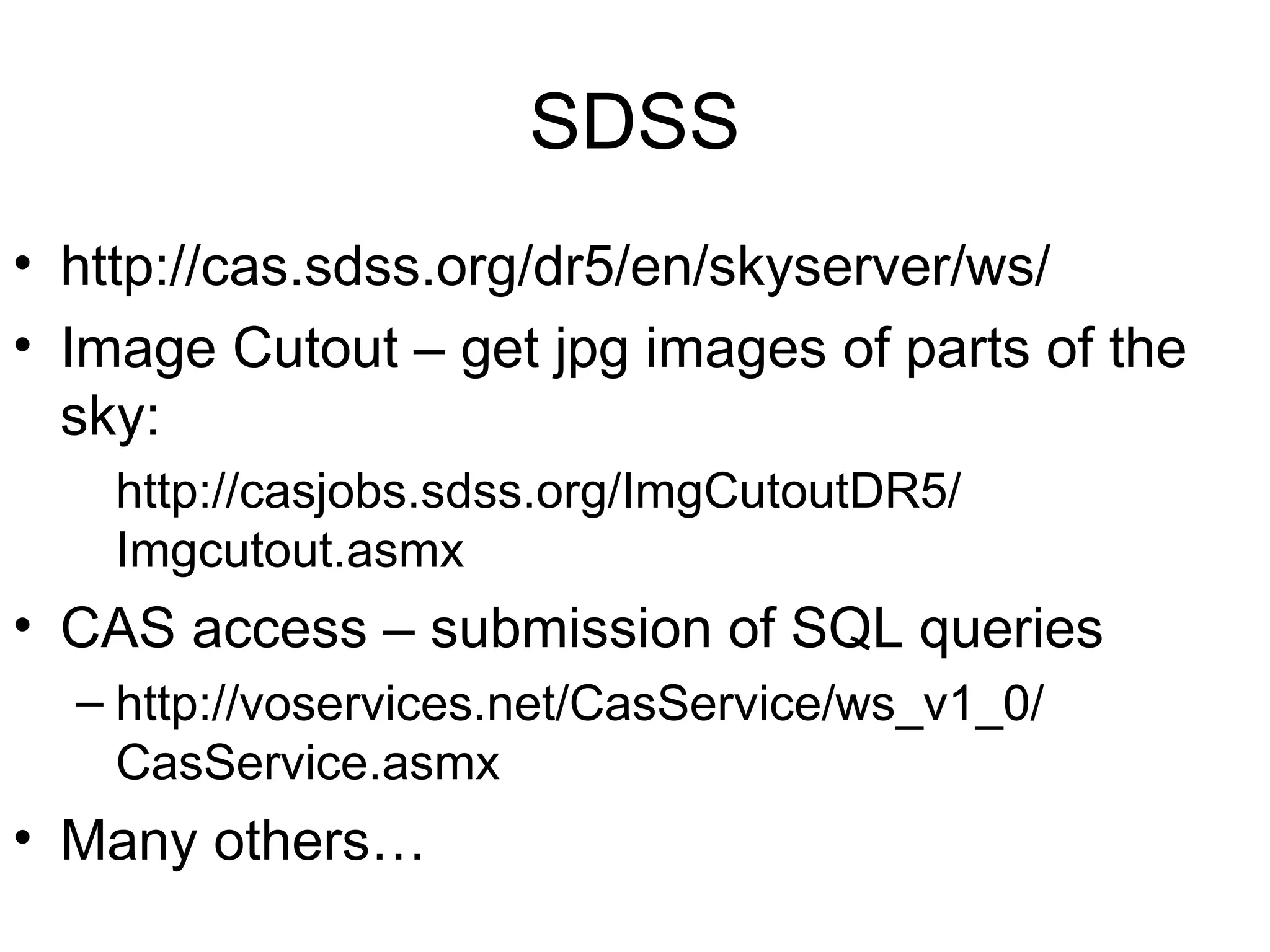
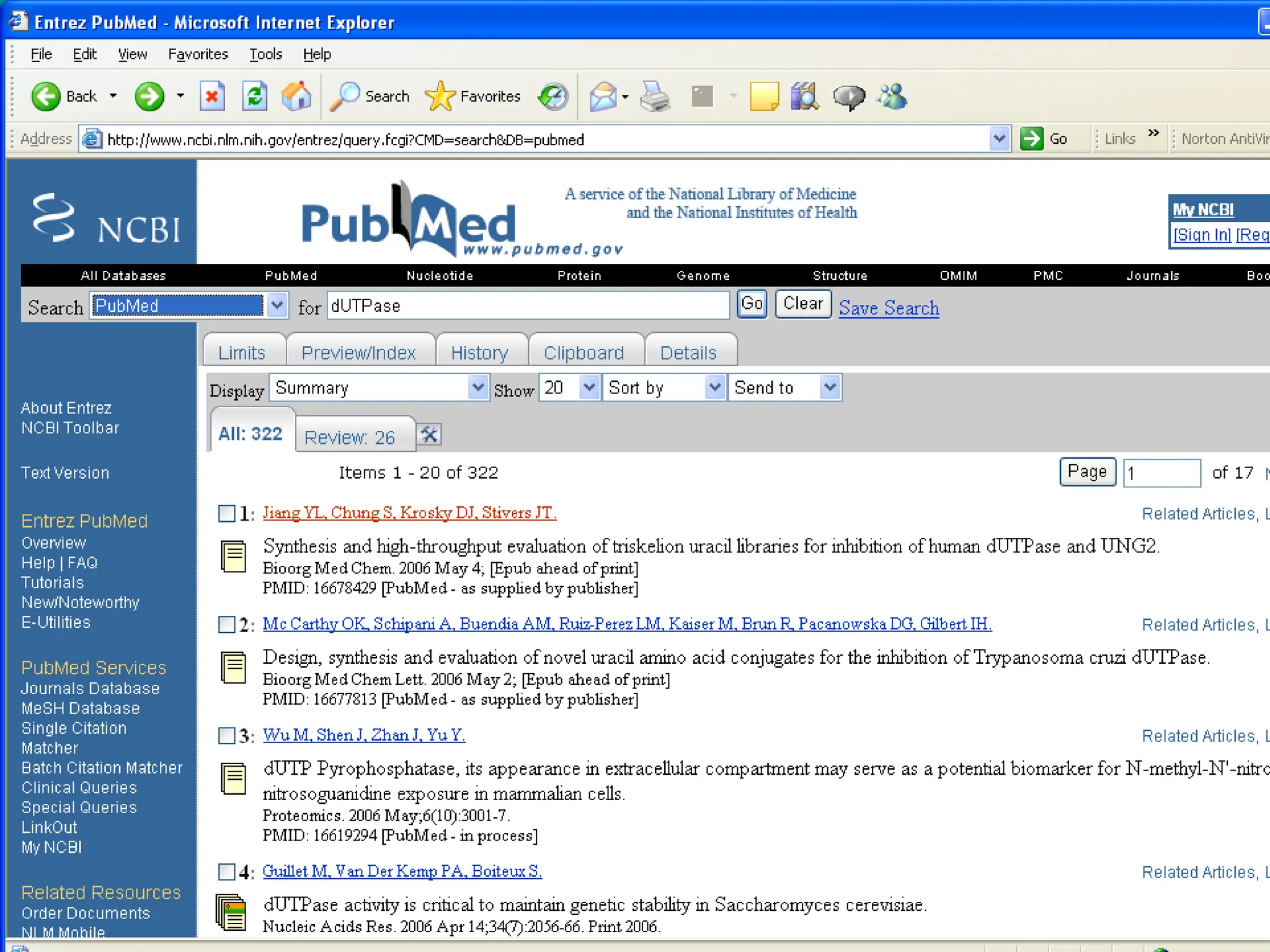
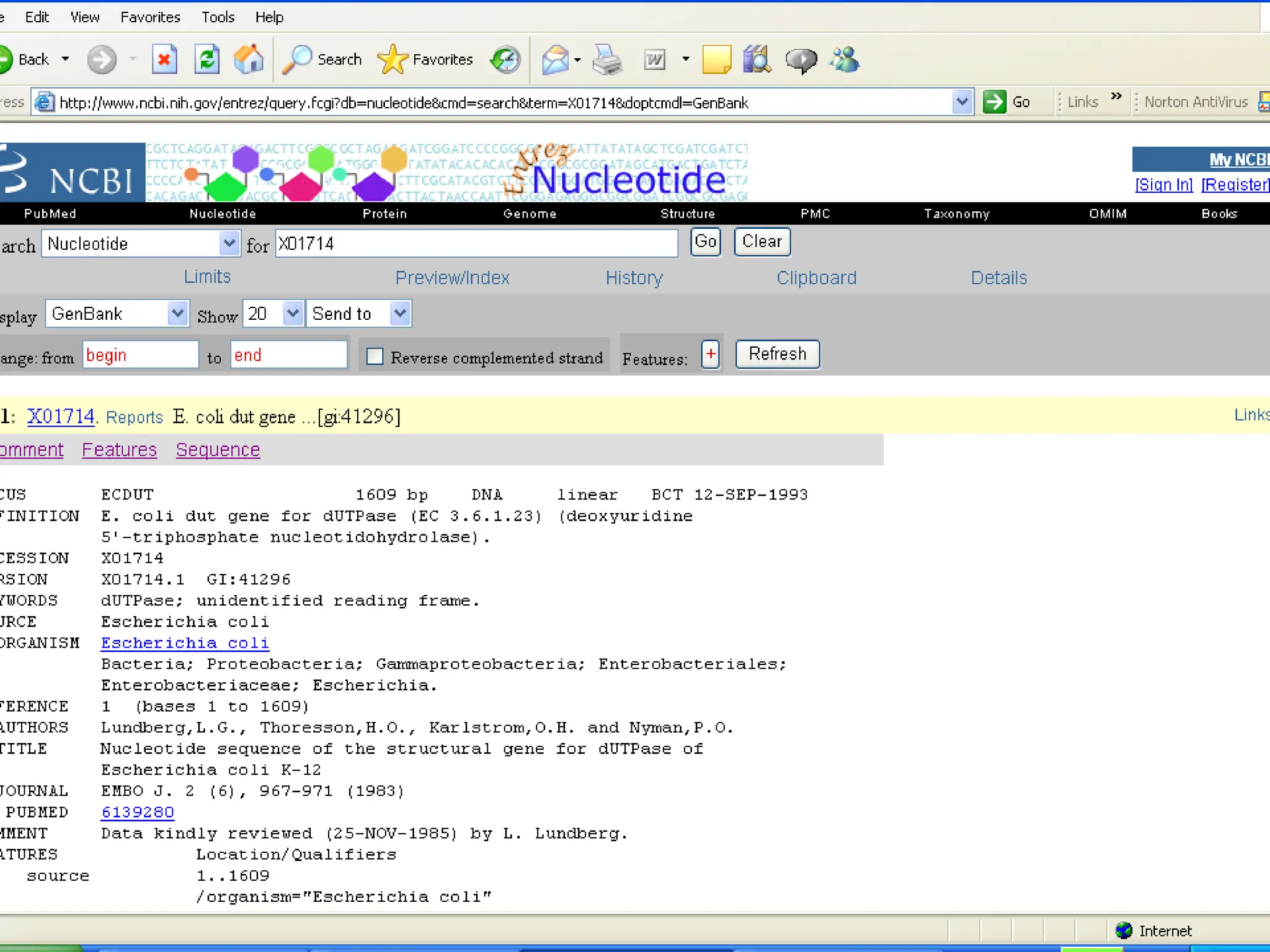
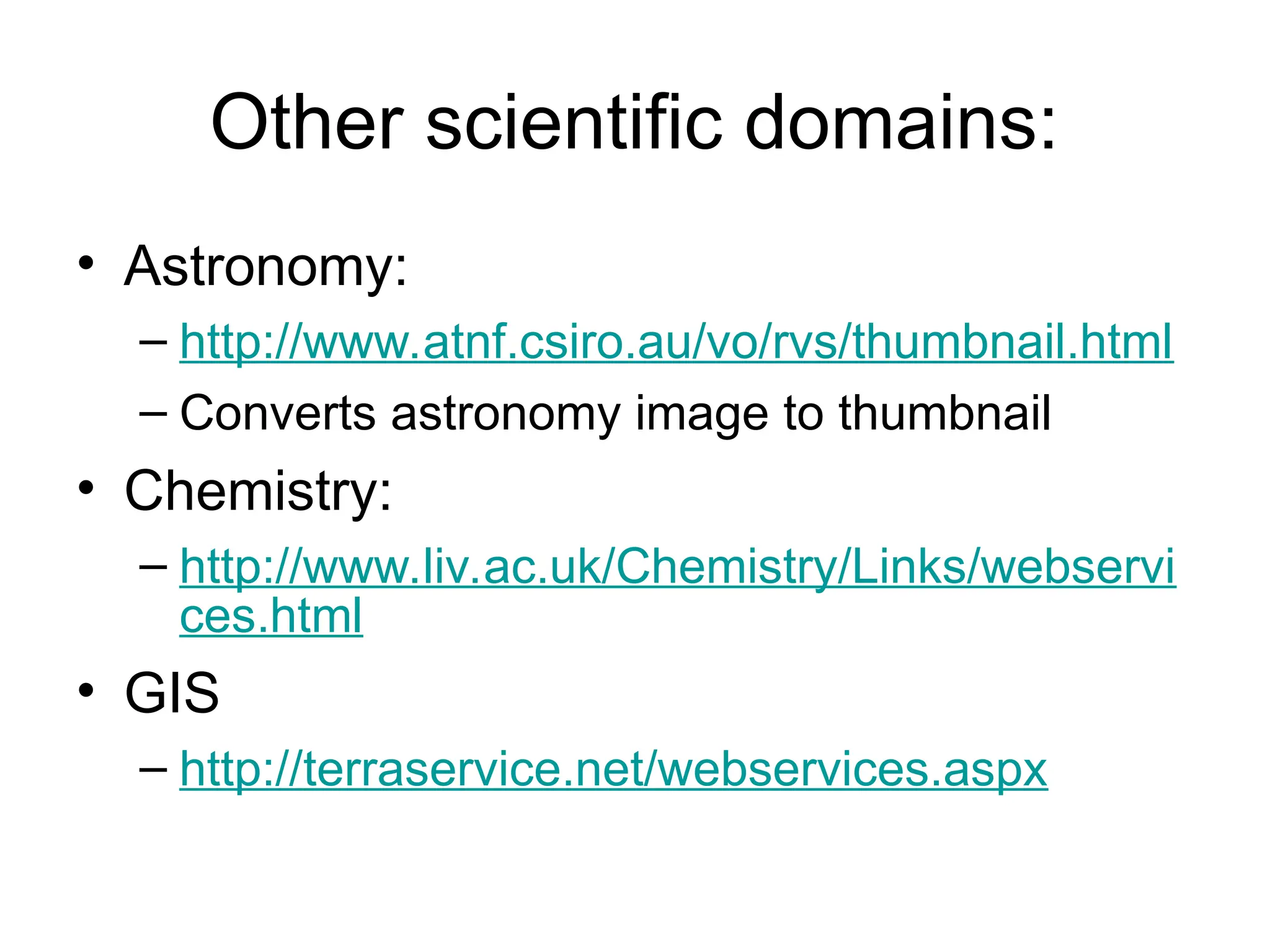
Web services are software accessible via the internet that use standardized messaging protocols for data exchange between applications. They enable tasks such as data retrieval and analysis through a service-oriented architecture leveraging established protocols like REST and SOAP. While offering significant advantages for scientific research, challenges such as interoperability, scalability, and quality control remain.


























![Example (continued) <portType name="Blast"> <operation name="searchSimple" parameterOrder="program database query"> <documentation>Execute Blast</documentation> <input name="searchSimple1In" message="tns:searchSimple1In" /> <output name="searchSimple1Out" message="tns:searchSimple1Out" /> </operation> ….[other operations] </portType>](https://image.slidesharecdn.com/websvcs-240925155037-4c09b243/75/Introduction-to-Web-Services-Architecture-33-2048.jpg)


![Using SOAP:Lite sub Blast_SOAP{ local $query_file = @_[0]; $service = SOAP::Lite -> service('http://xml.nig.ac.jp/wsdl/Blast.wsdl'); $service->proxy('http://localhost/', timeout => 60*60); return $service->searchSimple("blastn", “ddbjhum",$query_file); } $service connects to Blast web service at DDBJ site blastn is program, ddbjhum is database, $query_file contains sequence](https://image.slidesharecdn.com/websvcs-240925155037-4c09b243/75/Introduction-to-Web-Services-Architecture-36-2048.jpg)