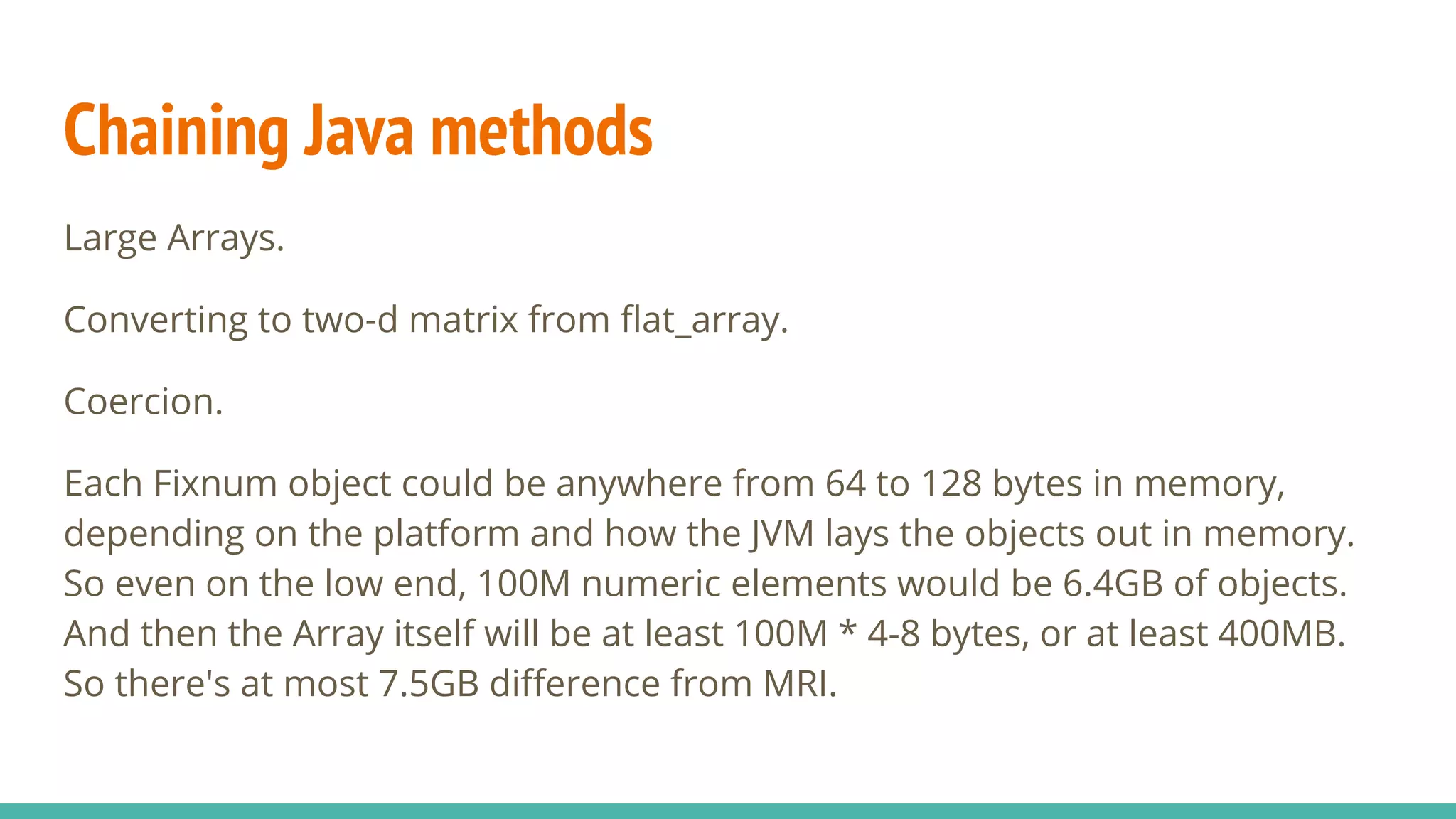
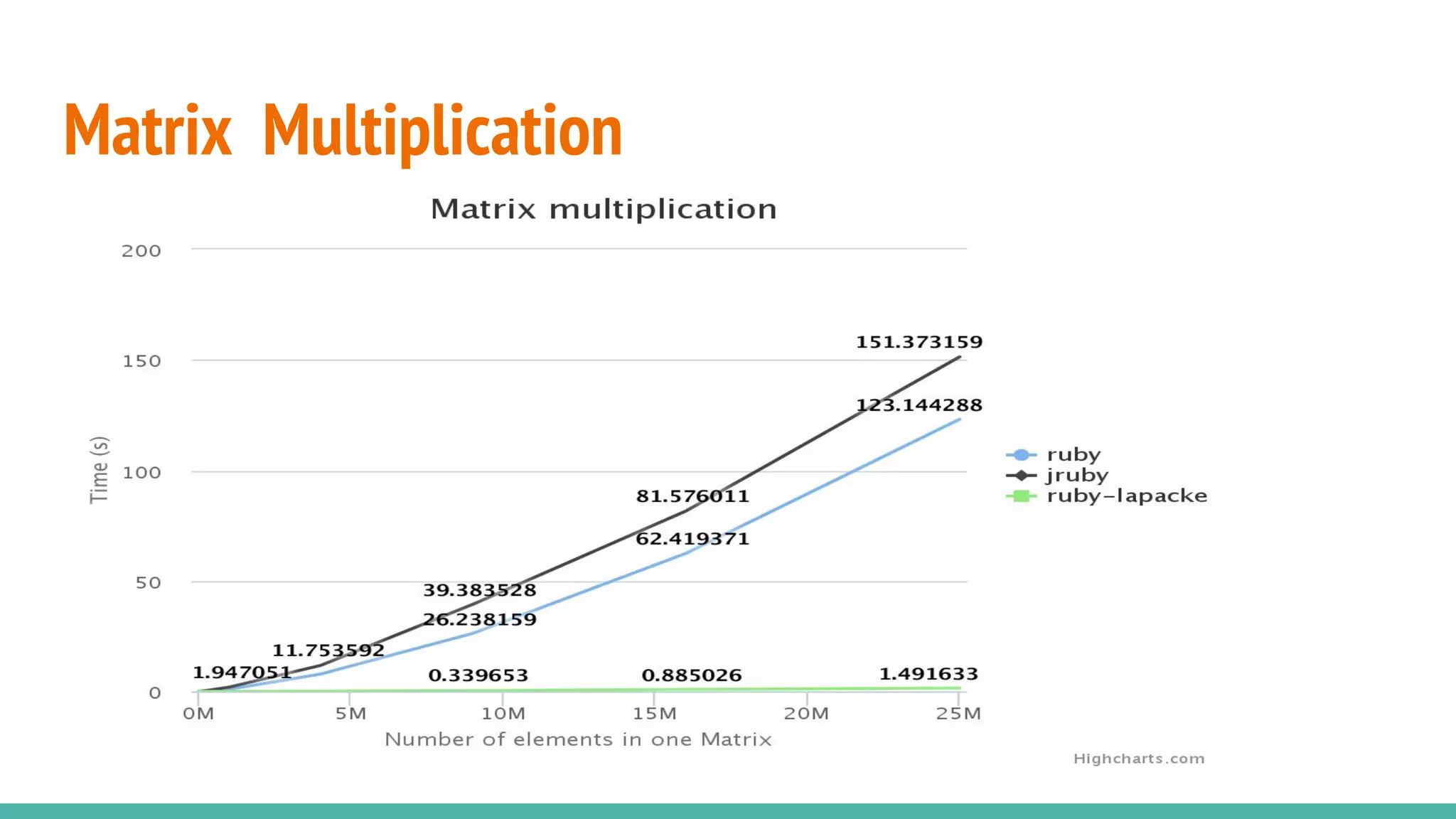
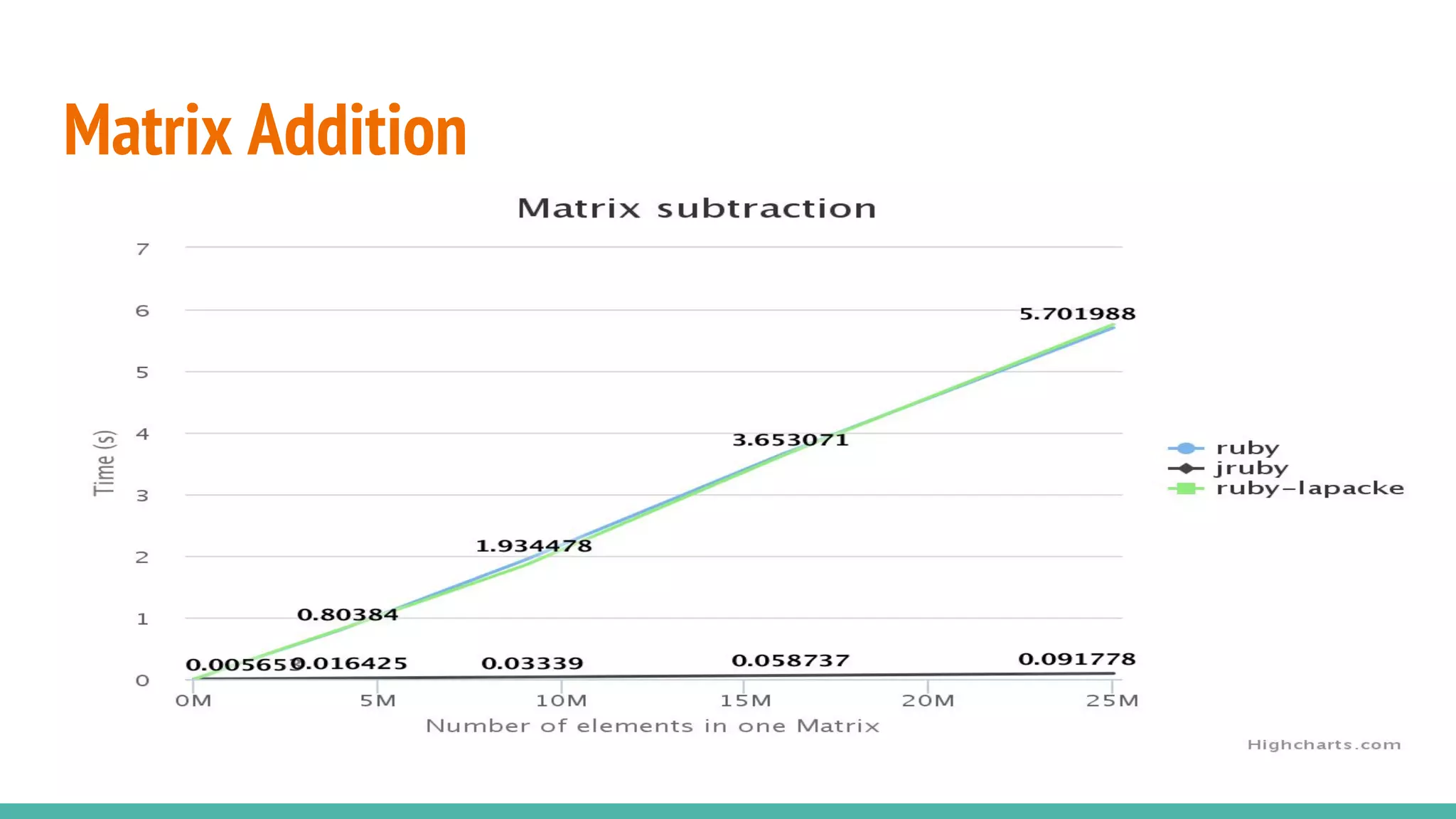
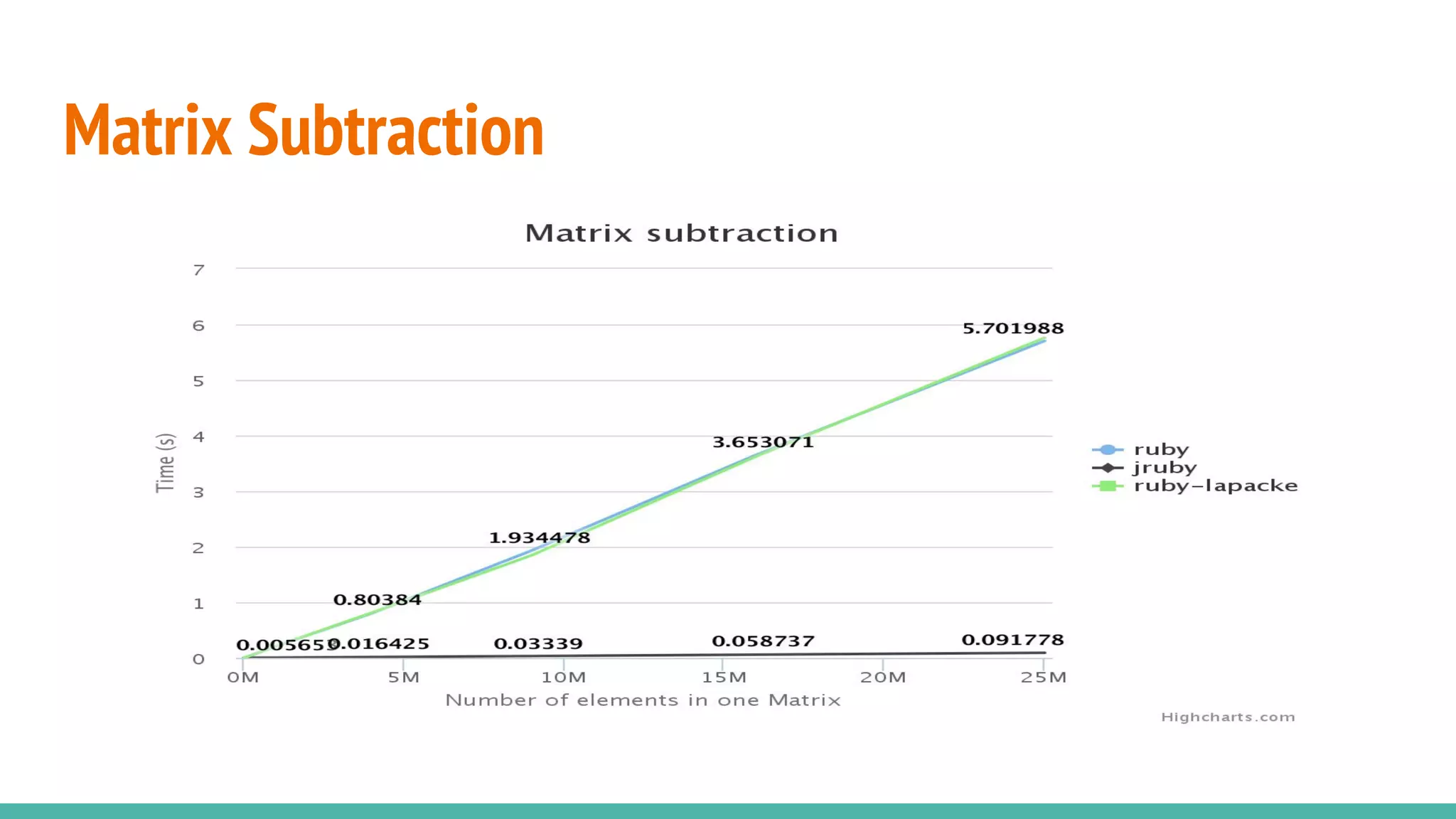
NMatrix is Sciruby's numerical matrix core that supports dense and sparse matrices on JRuby using Apache Commons Math. It allows creation and printing of matrices in Ruby and chaining of Java methods for optimization. Benchmarks show significant speed improvements when performing operations like copying a large matrix from one array to another compared to MRI Ruby. Memory management is improved through techniques like avoiding overcopying and type guessing. Mixed models for statistical analysis have also been ported to work with JRuby. Basic linear algebra operations like matrix multiplication, addition and subtraction are demonstrated.


![y = NMatrix.new([2,3],[1,2,3,4,5,6], dtype: :float32, stype: :dense) pp y [ [1,2,3] [4,5,6] ]](https://image.slidesharecdn.com/deccanrubyconf3-160806112615/75/Scientific-Computation-on-JRuby-4-2048.jpg)

![Benchmark b = Java::double[15_000,15_000].new c = Java::double[15_000,15_000].new # b= Array.new(25000000) index=0 puts Benchmark.measure{ (0...15000).each do |i| (0...15000).each do |j| b[i][j] = index index+=1 end end } index =0 puts Benchmark.measure{ (0...15000).each do |i| (0...15000).each do |j| c[i][j] = b[i][j] index+=1 end end }](https://image.slidesharecdn.com/deccanrubyconf3-160806112615/75/Scientific-Computation-on-JRuby-6-2048.jpg)